Small Molecule Alignment
Ligand-based screening is a critical component of computer-aided drug discovery (CADD) strategies for preclinical drug discovery. As a key member of ligand-based screening approaches, small molecule flexible alignment has been widely exploited to evaluate putative ligands. Working in the field of drug discovery for years, Creative Biolabs has established an advanced platform for small molecule alignment. Our scientists are dedicated to helping our clients determine the most likely ligand binding-pose.
How Does Small Molecule Alignment Work?
In drug discovery projects, atomic-level details of the structures of pharmaceutically relevant receptors are not available. In such cases, 3D alignment (or superposition) of putative ligands have been used to deduce structural requirements for biological activity. Until now, numerous methods are proposed for small molecular superposition or structural alignment. These methods can be classified largely into two types, namely point-based and property-based methods. In point-based methods, pairs of atoms or pharmacophoric points are superposed by the least-squares fitting. However, in property-based methods, various molecular properties are utilized for superposition, including electron density, molecular volume or shape, charge distribution or molecular electrostatic potential (MEP), hydrophobicity, hydrogen bonding capability, and so on.
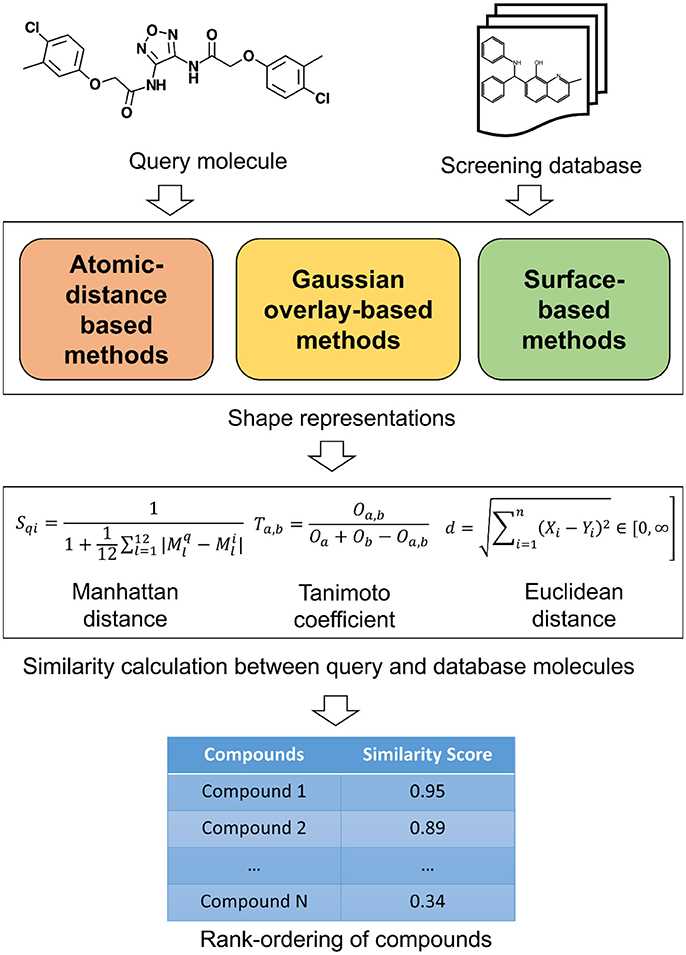 Fig.1 A schematic overview of similarity calculation between a query and database molecules. (Kumar, 2018)
Fig.1 A schematic overview of similarity calculation between a query and database molecules. (Kumar, 2018)
Generally, the 3D methods for aligning small molecules involve three basic components: (a) a descriptor to represent the molecule, (b) a scoring metric to rank molecule superimpositions based on the degree to which chemically similar functional groups, and (c) an optimization procedure to find the best possible alignment with respect to the chosen scoring function. For small-molecule alignment quality assessment, two main types of scoring functions are established. The first type is atom-based. When two molecules are being aligned, the score consists of a sum of terms that are based on intermolecular atom pairs (i.e., each pair has one atom coming from each molecule). The second type of score is field-based. The electrostatic or steric fields of the molecules or their surfaces are compared to arrive at a score.
Small Molecule Alignment Available at Creative Biolabs
With years of experience in virtual screening, Creative Biolabs has successfully established an advanced platform for small molecule alignment. Particularly, we have developed several property-based molecular alignment programs for flexible small molecule alignment. Aided by our top technology and professional scientists, we are able to rank each alignment with a score that indicates the quality of the alignment both in terms of internal strain and overlap of molecular features.
Features of Our Services
- Both standard and customized study proposal according to clients’ requirements
- Relative short delivery time
- Advanced technology platform and professional data analysis scientists
- Superior quality but competitive price
With the capabilities of various virtual screening methods, Creative Biolabs has established a powerful advanced platform that can be used to promote your drug discovery programs. We are committed to offering this high-quality screening assay to facilitate potential drug candidate discovery. Please feel free to contact us and let us know your demands, our team will get back to you as soon as we can.
Reference
- Kumar, A. and Zhang, K.Y. Advances in the Development of Shape Similarity Methods and Their Application in Drug Discovery. Frontiers in chemistry. 2018, 6: 315.
For Research Use Only.
