Assays using gene arrays are powerful tools for the parallel, high throughput detection, and quantification of many nucleic acid sequences. The use of gene array-based assays has quickly expanded in various fields of clinical diagnostics. As an industry-leading biotechnology company, Creative Biolabs has established an advanced technology platform with excellent experts, thus we are capable of providing gene array development services to support the progress of disease diagnostics strategies.
Introduction of Gene Array
Most microarray-based gene expression studies in humans have searched for genes that are differentially expressed in various pathologic states. Future clinical intervention, in the case of cancer, could be guided by diagnostic gene expression profiling. In general, DNA microarray hybridization applications are usually directed at gene expression analysis or for point mutation/SNP analysis. In addition to these important molecular biological and genomic research applications, microarray systems are also used for pharmacogenomic research, infectious and genetic disease and cancer diagnostics, and for forensic and genetic identification purposes. In addition, the implementation of new technologies such as array comparative genome hybridization (CGH) into the clinical practice has revolutionized the diagnostic workup in clinical practice and facilitated the identification of the molecular basis of many genetic diseases. First being developed as a research tool for the investigation of genomic imbalances in cancer, array CGH has become an essential and routine diagnostic tool replacing conventional cytogenetic methods and has been recognized as a first-tier clinical diagnostic test for patients with developmental disabilities or congenital anomalies
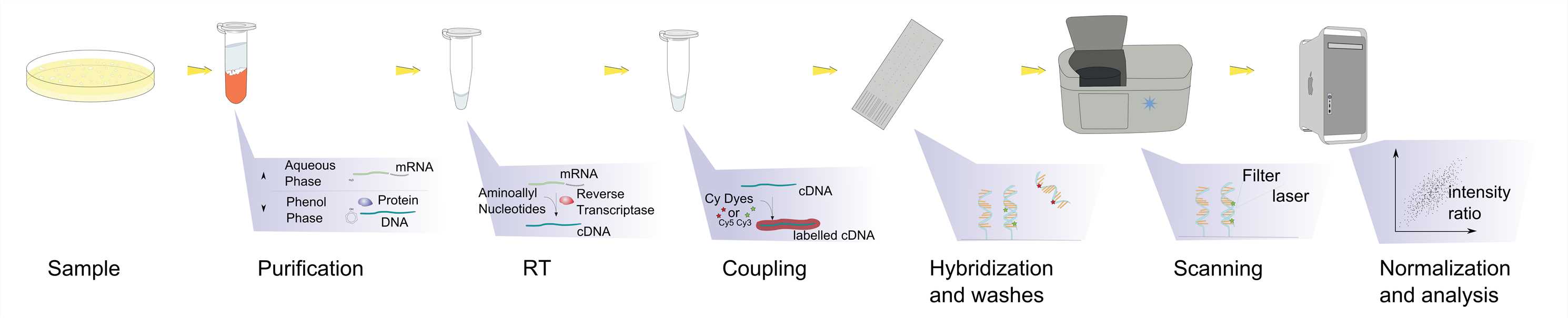 Fig.1 Microarray experiment workflow. Distributed under Public Domain, from Wiki, without modification.
Fig.1 Microarray experiment workflow. Distributed under Public Domain, from Wiki, without modification.
Examples of Gene Array Application for Disease Diagnostics
- DNA microarray applied in POC
Integration of various steps of DNA microarray assay in a miniaturized, portable, and stand-alone lab-on-a-chip (LOC) device is a crucial requirement for a variety of applications, especially point-of-care (POC) diagnostics. Current microarray technologies use separate instruments for sample preparation, DNA hybridization, signal visualization, and data interpretation. Moreover, some of these components such as the fluorescent scanners used for signal visualization are bulky instruments that are only available in well-equipped laboratories. Miniaturization of the microarray spots alleviates the need for fluorescent scanning and so no bulky fluorescent scanners are required. More importantly, with the aid of a Microfluidic network, all steps of the microarray test can be integrated into a single miniaturized device.
- Gene array applied in prenatal diagnostics
Array CGH, as well as SNP arrays, has made a significant impact on the diagnosis of individuals with congenital and developmental abnormalities including dysmorphic features, an intellectual disability, developmental delay, or multiple congenital anomalies without an obvious syndromic pattern. With the help of a variety of array platforms, the detection rate for copy number changes among these patients has an average diagnostic yield of 12.2% as compared to ~3% for G-banded chromosome analysis. Even greater detection rates (17.1%) have been observed with high-density arrays among neonates with birth defects. As a result, the American College of Medical Genetics (ACMG) recommends that array CGH be the first-line cytogenetic diagnostic test for patients with an unexplained developmental delay or intellectual disability, autism spectrum disorders, or multiple congenital anomalies.
Services at Creative Biolabs
Focusing on in vitro diagnostics (IVD) research over years, Creative Biolabs has established our technology platform with advanced facilities, the latest methods, and excellent experts. With the comprehensive platform we equipped step-by-step during practice, we are capable of providing high-quality gene array development services for disease diagnostics.
- One-stop worry-free services
- Ph.D. level expert team
- Flexible services form
- Efficient and cost-effective
Creative Biolabs has strong foundations and mature technologies. We are eager to offer our customers high-quality technology services. If you are interested in gene array development services or any other IVD development services, please don't hesitate to contact us for more information.
Published Data
1. Functional Gene Array for Bacterial Virulence Elements Detection
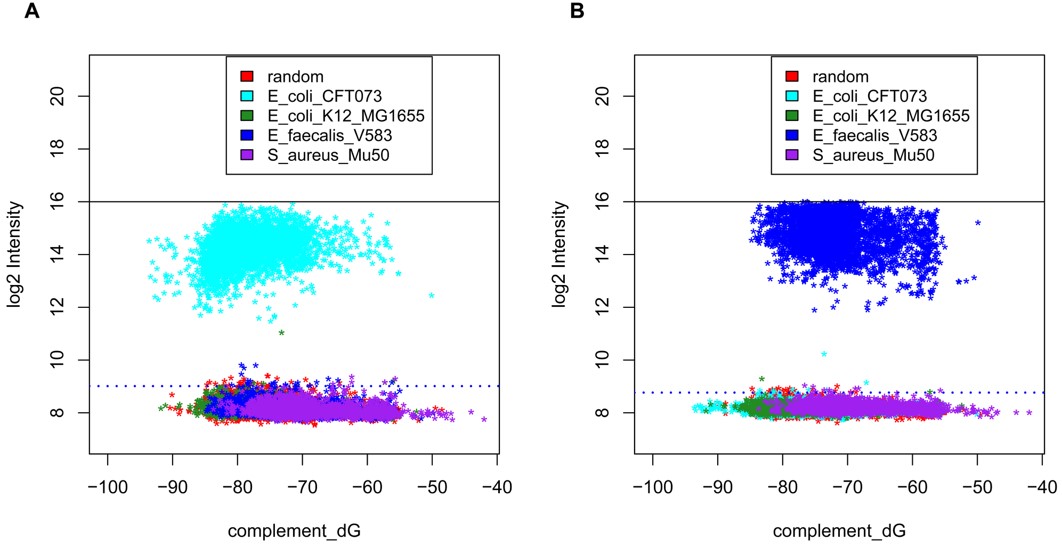 Fig.2 Target strain specificity of the functional gene arrays.1,3
Fig.2 Target strain specificity of the functional gene arrays.1,3
In this study, researchers developed high-density functional gene arrays to identify mechanisms related to virulence and antibiotic resistance. The first-generation array targeted genes from Escherichia coli strains K12 and CFT073, Staphylococcus aureus and Enterococcus faecalis. They optimized probe design parameters for gene family detection and differentiation. The array successfully identified orthologs for most gene families in bacteria within the same taxonomic family when tested with organisms at varying phylogenetic distances. Using whole-genome amplification, the array identified femtogram levels of DNA, whether spiked into an aerosol sample or from combinations of the four target organisms. This is the first report of a high-density microarray system aimed at detecting virulence gene families and genes specific to particular biothreat agents, offering valuable insights into the pathogenic potential and drug resistance profiles of unknown organisms in environmental samples.
2. Picoliter Well Array Chip-Based Digital Recombinase Polymerase Amplification for Absolute Nucleic Acid Quantification
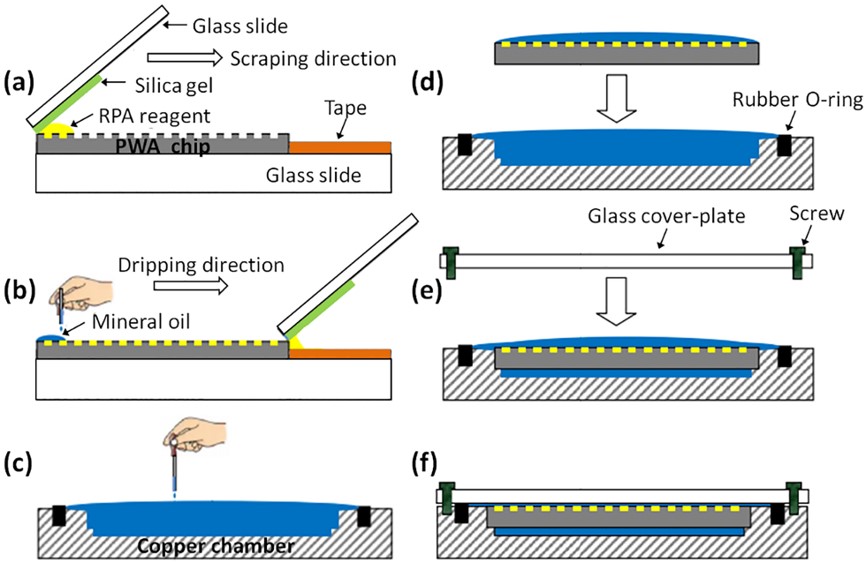 Fig.3 Workflow of sample loading and chip packaging.2,3
Fig.3 Workflow of sample loading and chip packaging.2,3
This study designed a picoliter well array (PWA) chip containing 27,000 uniform picoliter reactions (314 pL) for isothermal DNA quantification through digital RPA (dRPA) at 39°C. The sample loading method, employing a scraping liquid blade, was simple, rapid, and consumed less than 20 μL of reagents. Surface passivation with methoxy-PEG-silane prevented cross-contamination during the dRPA process. An innovative optical design allowed wide-field fluorescence imaging for both end-point and real-time analysis of wells across a 6-cm² area, eliminating the need for image stitching. This method successfully quantified serial dilutions of Listeria monocytogenes gDNA with an average error under 11%. dRPA processing took less than 30 minutes, four times faster than dPCR. This technique offers a simple, sensitive method for nucleic acid quantification in resource-limited settings and clinical diagnostics.
References
- Jaing, Crystal, et al. "A functional gene array for detection of bacterial virulence elements." PLOS one 3.5 (2008): e2163.
- Li, Zhao, et al. "Picoliter well array chip-based digital recombinase polymerase amplification for absolute quantification of nucleic acids." PLoS One 11.4 (2016): e0153359.
- Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.