What is Chromatin Immunoprecipitation (ChIP) Sequencing?
ChIP-sequencing, a potent technique that integrates ChIP assays with sequencing, allows for the identification of genome-wide DNA binding sites of proteins. ChIP sequencing serves as an indispensable tool, facilitating the precise identification and quantification of specific DNA sequences where proteins are bound or epigenetic modifications occur. Therefore, it has played a pivotal role in various applications, such as studies on gene regulation, transcription complex assembly, DNA repair, and developmental mechanisms.
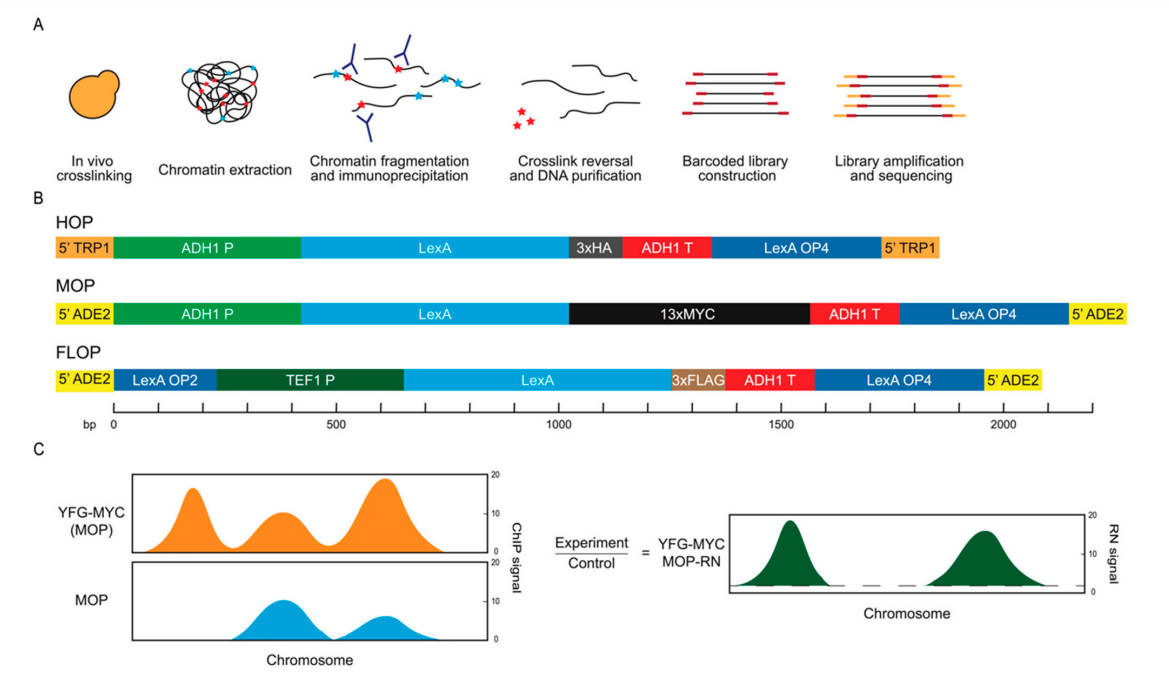 Fig.1 Control constructs and analysis scheme.1
Fig.1 Control constructs and analysis scheme.1
Principle of ChIP Sequencing
ChIP sequencing entails the enrichment of DNA fragments bound to the protein of interest using the ChIP technique, which is then followed by purification and the construction of a sequencing library. Afterward, these enriched DNA fragments undergo high-throughput sequencing. Researchers meticulously map the millions of sequence tags acquired onto the genome, offering profound insights into the DNA segments that engage with histones, transcription factors, and various other proteins across the entire genomic landscape.
Unique Features of ChIP Sequencing
- Reliability
Unlike arrays and other methods, ChIP sequencing distinguishes itself by eliminating the need for prior knowledge or probes derived from known sequences. This eliminates bias and errors, resulting in more reliable results.
- High Resolution
ChIP sequencing provides a more precise mapping of protein-DNA interactions throughout the entire genome, thereby enabling the accurate identification of protein binding sites, including histone modifications.
- High Precision
ChIP sequencing surpasses the constraints of fixed probe sequences employed in arrays, offering superior genome coverage, including repetitive regions frequently neglected by array-based methods.
- Comprehensive Analysis
Utilizing widely acknowledged software and cutting-edge programs, ChIP sequencing delivers a comprehensive analysis encompassing motif prediction, peak annotation, functional interpretation, and intuitive data visualization.
Applications
- Transcriptional Regulation
Identify transcription factor binding sites and study their role in gene activation or repression.
- Epigenetic Markers
Investigate the distribution and function of histone modifications associated with gene silencing, activation, or chromatin accessibility.
- Disease Mechanisms
Identify disruptions within regulatory networks that contribute to the onset and pathology of diseases.
- Drug Discovery
Evaluate the impact of therapeutic compounds on protein-DNA interactions, guiding the design of novel treatments.
- Enhancer and Promoter Mapping
Define regulatory elements that control gene expression programs in specific cell types or conditions.
ChIP Sequencing Service Workflow
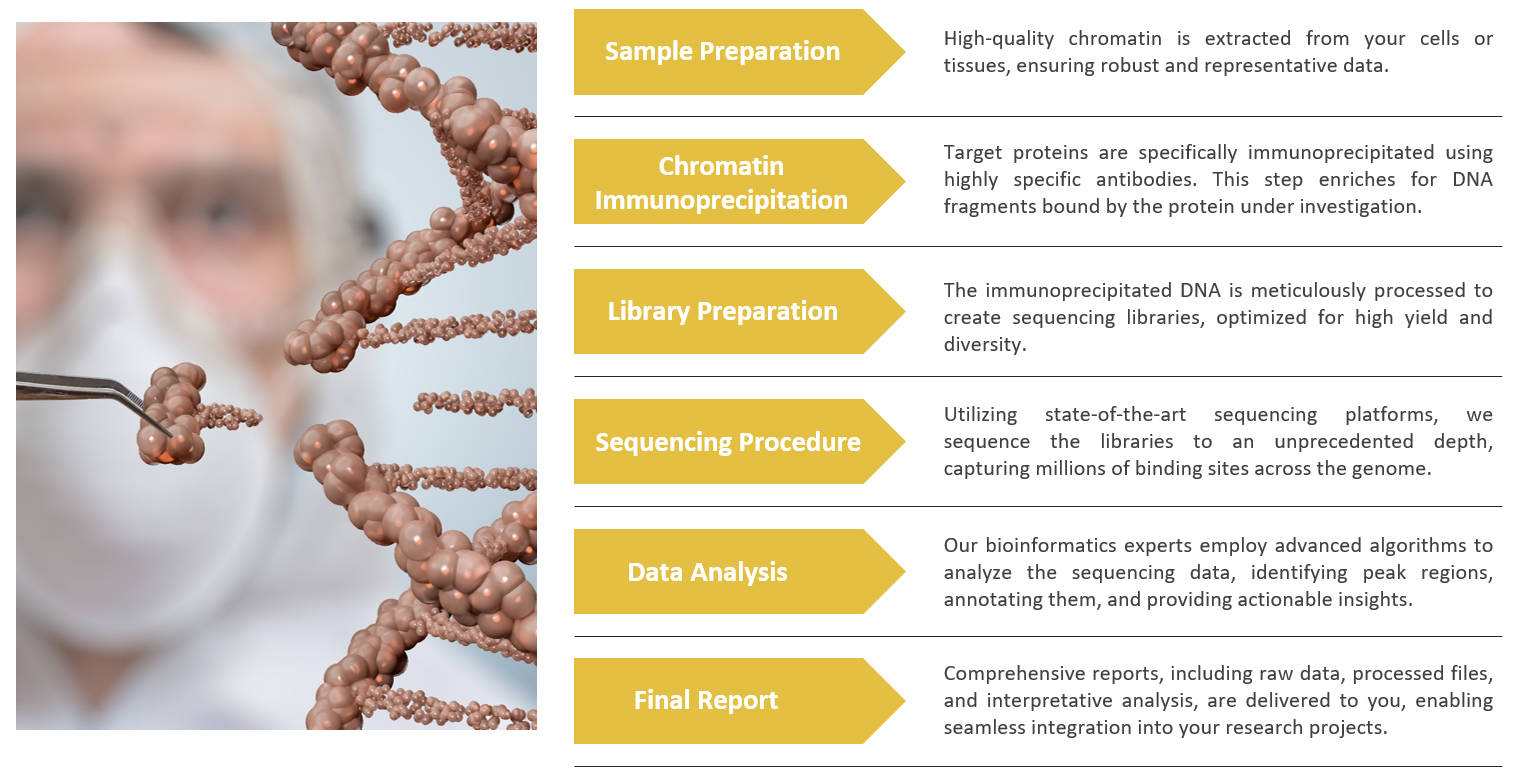
Delivery Results
- Related experimental results raw data
- Experimental report
- Data analysis
- Image and result analysis
- Bioinformatics analysis results
- Details in ChIP-sequencing
FAQs
Q: What are common sources of variability in ChIP-sequencing experiments?
A: Sources of variability can include cell culture conditions, fixation efficiency, chromatin shearing quality, antibody specificity and affinity, immunoprecipitation conditions, and sequencing depth. Rigorous controls, such as input DNA and IgG controls, are essential to mitigate these variables.
Q: What are the guidelines for delivering samples?
A: For ChIP-sequencing, the guidelines specify that the samples should be enriched DNA with a minimum concentration of 2 ng/μL and fragment sizes ranging from 100 bp to 500 bp.
Q: Can ChIP sequencing be used in combination with other sequencing technologies?
A: Yes, ChIP sequencing can be paired with RNA-Seq and whole-genome bisulfite sequencing to offer a comprehensive view of gene regulation and DNA methylation patterns.
At Creative Biolabs, our commitment to excellence, combined with our cutting-edge technology and personalized support, ensures that your research will benefit from the most accurate, comprehensive, and actionable data available. Contact us today to discuss your project.
Reference
- Petrie, Meghan V., et al. "Broadly Applicable Control Approaches Improve Accuracy of ChIP-Seq Data." International Journal of Molecular Sciences 24.11 (2023): 9271. Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.