Creative Biolabs is a leading global CRO company that focuses on in vitro diagnostics (IVD) for disease diagnosis. With our extensive experience and advanced platform, we have won a good reputation among our worldwide customers for accomplishing numerous challenging projects in this field. In our company, our scientists specialized in microsphere-based protein assay design and generation will work with you to develop the most appropriate strategy that will offer reproducible data for your research.
Introduction to Liquid Arrays for Protein Analysis
Recently, liquid arrays have been considered powerful tools for high-throughput protein analysis and drug candidate identification. In principle, a microsphere-based liquid array is capable of simultaneously detecting multiple protein types in a single system. Furthermore, this array allows detecting different kinds of samples with extremely low amounts. The arrays are usually designed by immobilizing proteins into certain slides. Meanwhile, they will be probed by a wide battery of fluorescent tags or radioisotope labels for detecting a range of protein functions or protein activities. As a consequence, these microsphere-based liquid arrays have been divided into two main subtypes, analytical microsphere-based protein assays, and functional microsphere-based protein assays. Up to now, many efforts have been made to improve their sensitivity, specificity, and accuracy in a broader field of protein studies, including protein-protein interaction, biochemical activity, protein binding, as well as immune responses.
Microsphere-based Protein Assays
With the commitment of being your best drug development partner, Creative Biolabs worked on a wide range of protein detection and analysis services based on various liquid arrays. We utilize unrivaled, proprietary probe selection, solid-surface support design, array optimization, advanced project lifecycle management, as well as real-time data analysis to ensure the ideal outcomes. Till now, we have established a panel of microsphere-based protein assays to reveal various biochemical properties of proteins, such as protein-protein binding activities, protein-DNA binding activities, protein-drug interaction, and protein expression profiling. For example, a collection of high-throughput, sensitive, specific microsphere-based liquid chips have been designed and synthesized for detection and identification of respiratory viruses, such as human papillomavirus (HPV).
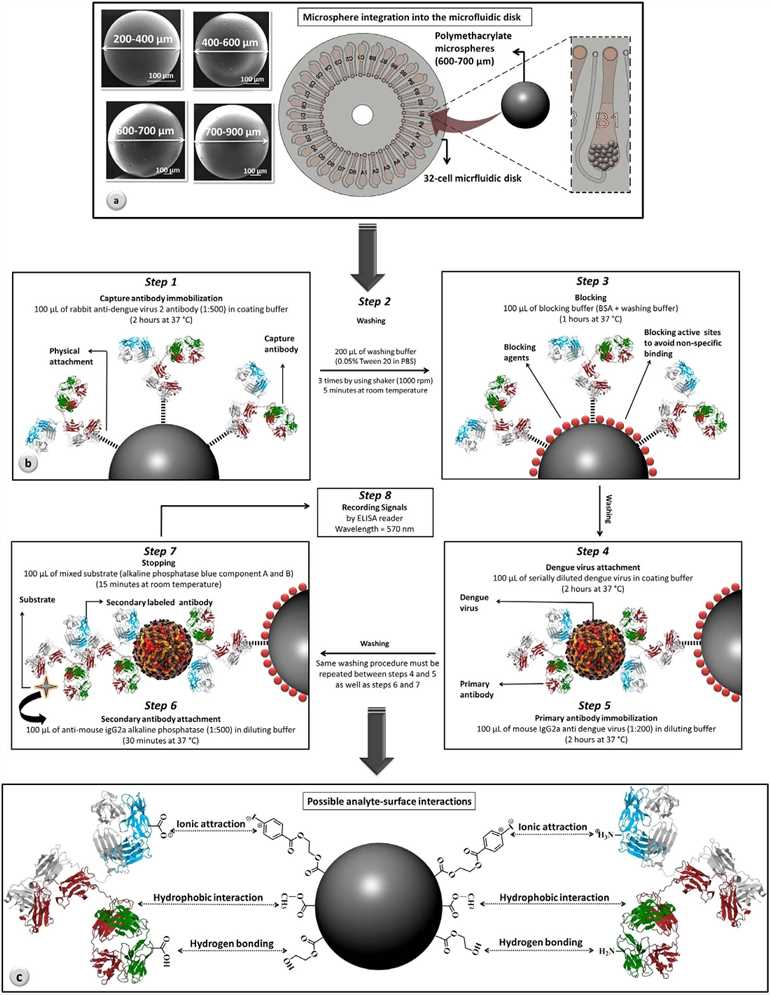 Fig.1 Microfluidic disk with integrated microspheres.1, 2
Fig.1 Microfluidic disk with integrated microspheres.1, 2
Furthermore, a panel of antibody array-based assays, like affinity testing and candidate screening assay, have been used for determining multiple protein targets simultaneously with a fluorescence readout. In Creative Biolabs, our microsphere-based antibody array technology employed here combines the advantages of fluorescence microscope array analysis and suspension array based on microspheres. Chemically etched high-density fiber bundles can produce a large number of microwaves in which antibody functionalized microspheres are randomly deposited to form antibody arrays. In a recent study, antibody arrays can screen for simultaneously detecting more than 65 proteins with high specificity, femtomolar sensitivity, and minimal sample consumption. Also, the results have suggested that these microsphere-based antibody arrays should be an attractive alternative to commercially available quantitative ELISA kits for various disease diagnosis.
Through our full spectrum of early discovery, nonclinical, and commercialization services, Creative Biolabs is committed to developing the most promising liquid array diagnostics tools for our clients. We have completed a series of challenges in the past years. In particular, we have established a well-mature microsphere-based protein assays development platform which enables us to offer a series of high-quality products for diagnosing diseases. If you are interested in our services, please contact us or send us an inquiry.
Published Data
1. A New Microsphere-Based Antibody Array Analysis of Primary Human Keratinocyte and Leukocyte Proteins
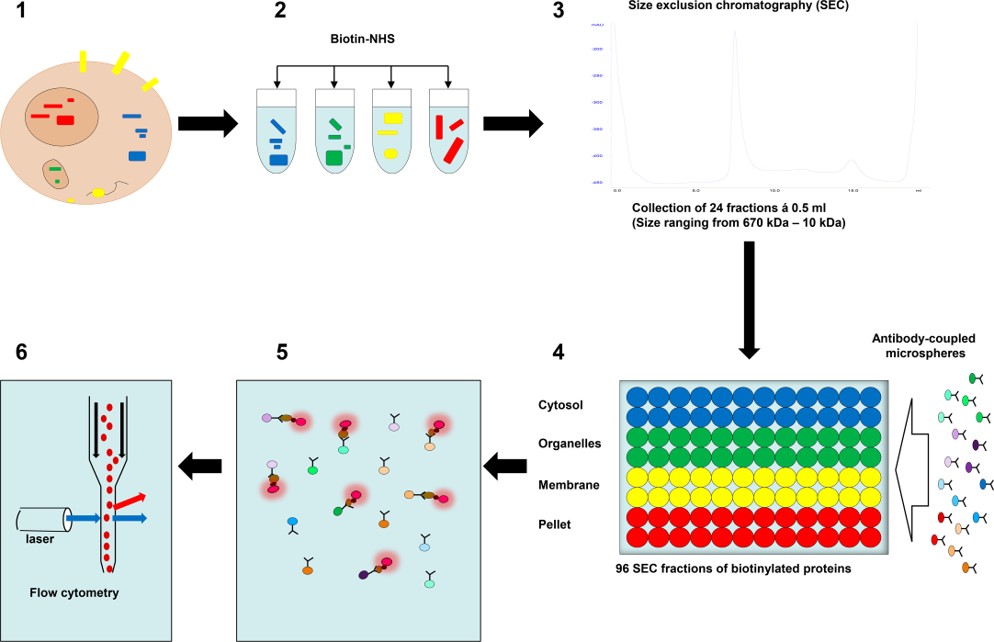 Fig.2 Schematic representation of microsphere affinity proteomics.3,2
Fig.2 Schematic representation of microsphere affinity proteomics.3,2
This study utilized differential detergent fractionation followed by size exclusion chromatography (SEC) to separate biotinylated proteins from human leukocytes and primary keratinocytes in a high resolution. A total of 96 sample fractions from each cell type were assessed using microsphere-based antibody arrays in conjunction with flow cytometry, a method referred to as microsphere affinity proteomics (MAP). This approach resolved monomeric proteins and multi-molecular complexes in various cellular compartments into distinct antibody reactivity peaks across fractions. The specificity of these peaks was validated by matching their position with known information on subcellular location, protein size, and cell type-specific protein expression. Reactivity patterns from different antibodies to the same or similar proteins provided additional supporting evidence. The high-resolution MAP technique addresses challenges in antibody specificity and protein labeling, enabling large-scale protein analysis and advancing cell biology research, particularly in dermatology and antibody validation.
2. High-Resolution Antibody Array-Based Proteomic Analysis for Childhood Acute Leukemia Cells
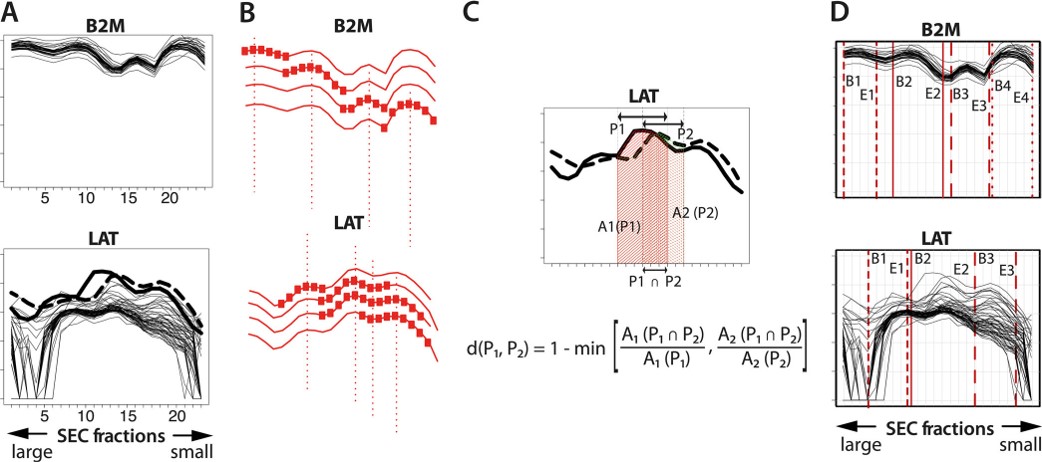 Fig.3 SEC-MAP technology for leukemia immunophenotyping.4,2
Fig.3 SEC-MAP technology for leukemia immunophenotyping.4,2
In this study, researchers used SEC-Microsphere-based Affinity Proteomics (SEC-MAP), a novel microsphere-based antibody array platform, to detect hundreds of proteins from a single sample. They also created semi-automated bioinformatics tools to tailor this technology for high-throughput proteomic screening in pediatric acute leukemia patients. To validate SEC-MAP for leukemia immunophenotyping, they tested 31 diagnostic markers in parallel via flow cytometry and SEC-MAP and found 28 antibodies suitable for both methods. Eighteen antibodies showed excellent quantitative correlation (p < 0.05). SEC-MAP was applied to 57 diagnostic samples from acute leukemia patients, using 632 antibodies to detect 501 targets. Among these, 47 targets showed differential expression across at least two leukemia subgroups. Overall, SEC-MAP proved to be a powerful tool for detecting proteins in clinical samples, revealing protein size and expression differences across leukemia subgroups.
References
- Hosseini, Samira, et al. "Microsphere integrated microfluidic disk: synergy of two techniques for rapid and ultrasensitive dengue detection." Scientific reports 5.1 (2015): 16485.
- Distributed under Open Access license CC BY 4.0, without modification.
- de la Rosa Carrillo, Daniel, et al. "High-resolution antibody array analysis of proteins from primary human keratinocytes and leukocytes." Plos one 13.12 (2018): e0209271.
- Kanderova, Veronika, et al. "High-resolution antibody array analysis of childhood acute leukemia cells." Molecular & Cellular Proteomics 15.4 (2016): 1246-1261.
For Research Use Only.