Single strand conformation polymorphism (SSCP) analysis is a simple and sensitive method for gene typing and mutation detection. Creative Biolabs is skilled in the design and development of SSCP assays for disease-related DNA small mutation detection, providing a useful and effective tool for disease diagnosis.
Introduction of Single-Strand Conformation Polymorphism (SSCP) Analysis
SSCP is an electrophoretic technique that discovers new DNA polymorphisms apart from DNA sequencing. The principle of SSCP is that the electrophoretic mobility of a single-stranded DNA fragment is dependent on the secondary structure of the fragment. Altered conformation changes due to a single base change in the sequence cause single-stranded DNA to migrate in different ways under a non-denaturing gel. SSCP has been used to detect base substitutions, small deletions, or insertions in the DNA. The main advantage of the SSCP technique is the ability to detect mutations at a variety of positions in DNA fragments with good sensitivity. SSCP has been shown to detect single-base differences in 99% of DNA fragments with up to 300 bases in size. Now SSCP technique has been widely used as a diagnostic tool in molecular biology.
 Fig.1 Picture of an SSCP gel. Distributed under CC BY-SA 3.0, from Wiki, without modification.
Fig.1 Picture of an SSCP gel. Distributed under CC BY-SA 3.0, from Wiki, without modification.
Applications
Application of SSCP Analysis in the Diagnosis of Androgen Insensitivity Syndromes
Androgen insensitivity syndromes are a heterogeneous group of related disorders characterized by defective sexual differentiation in karyotypic males. Mutations in the androgen receptor (AR) gene have been proved an association with the occurrence of androgen insensitivity syndromes. A study used a combination of SSCP analysis and direct DNA sequencing to analyze mutations in the androgen receptor gene. Results showed that point mutations in the AR gene in all six patients were detected. SSCP-based AR gene mutation detection serves as a useful tool for the diagnosis of androgen insensitivity syndromes.
Application of SSCP Analysis in the Diagnosis of Duchenne Dystrophy
Duchenne dystrophy is the most common, lethal, X-chromosome-linked recessive disorder. This disease is caused by deficient some small mutations of the dystrophin gene. A study showed SSCP is a highly sensitive method for the detection of small mutations of the dystrophin gene. Using the SSCP technique, small mutations (such as nonsense mutations and microdeletions or insertions), or whole exon deletions in 73 (78.5%) of 93 patients with Duchenne dystrophy were detected. SSCP may increase the detection of small dystrophin gene mutations.
Our Capabilities
As an expert in the field of IVD development, Creative Biolabs is specialized in flexibly leveraging a variety of biological technologies to support the development of IVD assays to assist in disease diagnosis. Leveraging the SSCP analysis technique, we support the detection of a variety of known or novel small genomic variants such as substitutions, small deletions, and insertions, without using DNA sequencing, to facilitate the detection of genetic disorders. Our SSCP testing service package includes DNA extraction, prime design and PCR, sample preparation for SSCP analysis, capillary electrophoresis, and data analysis.
If you are interested in our services, please don't hesitate to contact us for more details. We are committed to becoming your reliable partner to develop novel specific IVD assays for disease diagnosis research.
Published Data
1. Validation of a Sensitive Fluorescent SSCP Assay for Detection of p53 Mutations
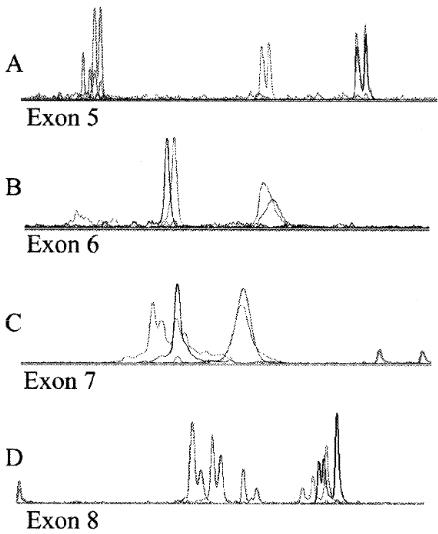 Fig.2 Fluorescent SSCP detection in cell lines.1
Fig.2 Fluorescent SSCP detection in cell lines.1
In this study, researchers developed a fluorescence-based SSCP method, providing rapid and sensitive detection of mutations in exons 5–8 of the human p53 gene. The method utilized a DNA sequencer to detect unique colors for each strand, along with precise lane alignment for better detection of mobility shifts. To validate the approach, 21 cell lines with known mutations in p53 exons 5–8 were analyzed by SSCP under various gel conditions. The sensitivity for mutation detection was 95% across all cell lines, with no false positives in 10 normal DNA samples for all four exons. Mixing known amounts of tumor and normal DNA showed that mutations could be detected even when tumor DNA was mixed with 80% normal DNA. Fluorescent SSCP analysis using the sequencer proves to be a valuable tool for screening large numbers of samples for p53 mutations in cancer research.
Reference
- Moore, L., et al. "Validation of fluorescent SSCP analysis for sensitive detection of p53 mutations." Biotechniques 28.5 (2000): 986-992. Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.