Escherichia coli-derived Exosome Research & Application
Overview Workflow Insights Our Advantages Client Perspectives FAQs
Escherichia coli (E. coli), one of the most well-studied Gram-negative bacteria, secretes nanosized extracellular vesicles known as exosomes or outer membrane vesicles (OMVs). These vesicles have become powerful models for studying microbial communication, host-pathogen interaction, and vesicle-mediated signaling.
At Creative Biolabs, we specialize in the isolation and characterization of E. coli-derived exosomes, enabling researchers to uncover their molecular roles in cell signaling, immune modulation, and intracellular communication. Our services are designed to support academic and biotechnology clients pursuing basic and translational exosome biology, without engaging in clinical or therapeutic applications.
Overview of E. coli-Derived Exosomes
Recent studies have shown that E. coli-derived exosomes serve as sophisticated mediators of bacterial–host communication. They carry virulence-associated biomolecules and regulatory RNAs that modulate cellular pathways, affecting both immune defense and cellular homeostasis. Key functional characteristics include:

Regulation of Autophagy
E. coli-derived exosomes can alter the host cell's autophagic process. Certain virulence factor-associated vesicles, such as those expressing HlyF, can block autophagic flux and prevent the degradation of intracellular bacteria.

Inflammatory Pathway Activation
These vesicles can trigger inflammatory responses, including activation of noncanonical inflammasome pathways, contributing to host tissue stress and immune signaling cascades.

Host–Bacteria Crosstalk Modeling
E. coli exosomes are ideal experimental tools for studying vesicle-driven communication, shedding light on microbial strategies for survival and persistence within the host environment.

Model for Gram-Negative Vesicle Biology
As the most accessible and well-characterized bacterial species, E. coli serves as a foundational system for establishing standard methods to study OMVs across different Gram-negative species.
Discuss with Creative Biolabs how we can support your E. coli vesicle research with proven technical depth and analytical precision.
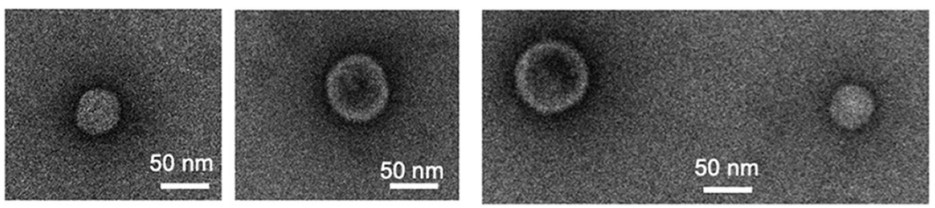 Fig.1 Transmission electron microscopy of Escherichia coli-derived exosomes.1
Fig.1 Transmission electron microscopy of Escherichia coli-derived exosomes.1
Standard Workflow for E. coli-Derived Exosome Preparation
Creative Biolabs offers a reliable, reproducible pipeline for isolating E. coli-derived exosomes suitable for experimental and analytical research.
1.Culture Preparation
-
Incubate E. coli in optimized liquid medium under shaking conditions for 8–10 hours.
-
Maintain culture temperature and aeration parameters for optimal vesicle release.
2.Primary Centrifugation
-
Centrifuge at low speed (4°C) to remove large bacterial cells and aggregates.
3.Filtration
-
Filter the resulting supernatant through sterile 0.22 μm filters to ensure removal of residual bacteria.
4.Concentration
-
Concentrate the filtered supernatant using ultrafiltration membranes to enrich extracellular vesicles.
5.Ultracentrifugation
-
Perform high-speed ultracentrifugation to pellet E. coli-derived exosomes.
-
Resuspend the vesicle pellet in sterile PBS buffer for downstream use.
Reach out to Creative Biolabs to design an optimized E. coli exosome isolation and analysis workflow tailored to your research goals.
Highlights from Research on E. coli-Derived Exosomes
Creative Biolabs continuously monitors advances in exosome-related microbiology to help researchers identify emerging directions and reproducible protocols. The following studies summarize key findings from independent research groups investigating E. coli-derived exosomes.
|
RESEARCH FOCUS
|
KEY FINDINGS AND INTERPRETATIONS
|
|
Influence of HlyF-Positive E. coli Exosomes on Autophagy
|
Exosomes derived from E. coli expressing the virulence factor HlyF significantly increased intracellular LC3-II accumulation and LC3-positive foci in a dose-dependent manner. Mutated HlyF strains did not induce this effect, confirming HlyF's critical role in modulating host autophagy.
|
|
Autophagosome Accumulation Mechanism
|
Co-localization studies using fluorescence microscopy and TEM revealed that LC3-positive vesicles induced by HlyF-positive exosomes possessed double membranes, confirming autophagosome identity. Phagocytosis inhibitors had no impact, indicating the autophagic rather than endocytic nature of vesicle formation.
|
|
Evidence of Autophagy Blockade
|
Blocking autophagic degradation with chloroquine did not further increase LC3 levels, suggesting HlyF-positive exosomes caused an autophagy blockade rather than activation. Fluorescent reporter cell analysis identified the defect at the autophagosome–lysosome fusion stage.
|
|
Inflammasome Activation
|
E. coli exosomes induced IL-1β secretion and macrophage cell death in a pattern consistent with noncanonical inflammasome activation. This response was significantly reduced in inflammasome-deficient models, supporting the hypothesis of vesicle-mediated inflammasome triggering.
|
|
Host–Bacteria Interaction Model
|
Combined analyses demonstrate that E. coli-derived exosomes act as potent mediators in host-pathogen interactions, providing an ideal in vitro and in vivo model for understanding vesicle-driven infection biology.
|
Collaborate with Creative Biolabs to develop controlled studies that replicate or expand on these global findings.
Advantages of Partnering with Creative Biolabs
Creative Biolabs has built a strong reputation in bacterial exosome studies through years of technical refinement and collaboration with microbiology research teams worldwide. Our advantages include:
 Proven Expertise:
Proven Expertise:
Deep understanding of vesicle biology across multiple Gram-negative species, including E. coli, Klebsiella, Haemophilus, and Moraxella.
 Customizable Services:
Customizable Services:
Modular service design allows clients to choose between basic vesicle isolation or expanded optional analyses (proteomics, RNA, imaging).
 Standardized Workflow:
Standardized Workflow:
Consistent yield and reproducibility across batches with stringent sterility and contamination control.
 Technical Flexibility:
Technical Flexibility:
Access to a wide array of analytical methods depending on the strain, growth conditions, and experimental objective.
 Scientific Integrity:
Scientific Integrity:
Transparent data handling, reproducible protocols, and comprehensive documentation for every project phase.
 Collaborative Approach:
Collaborative Approach:
Creative Biolabs scientists maintain direct communication throughout the study, ensuring that experimental parameters align with the client's hypotheses and publication standards.
Start your collaboration with Creative Biolabs and experience precision-driven bacterial exosome research support.
Voices from Our Customers
"Creative Biolabs' bacterial exosome services provided us with the cleanest E. coli vesicle preparations we've ever tested. Their centrifugation and filtration workflow produced reproducible results across multiple batches."
"We appreciated how Creative Biolabs helped us integrate optional proteomic profiling into our E. coli OMV study. Their data validation reports were thorough and transparent, making our publication process smooth."
"What sets Creative Biolabs apart is their commitment to collaboration. The scientists were responsive to every question, providing insightful feedback on vesicle quantification and autophagy assays."
Partner with Creative Biolabs to ensure consistency, accuracy, and insight in your bacterial vesicle studies.
Creative Biolabs provides customized research services for Escherichia coli-derived exosome isolation, profiling, and analysis. Our expertise in gram-negative bacterial vesicles helps researchers explore exosome-mediated signaling, autophagy regulation, and host-pathogen interactions with reliable data and reproducible workflows. Contact us to discuss your E. coli-derived exosome project with our expert team.
FAQs
Q: What are Escherichia coli-derived exosomes and how are they formed?
A: Escherichia coli-derived exosomes, often referred to as outer membrane vesicles (OMVs), are nanoscale lipid bilayer structures released from the outer membrane of E. coli. They are formed through the budding of the outer membrane, which allows the encapsulation of various biomolecules.
Q: What functional roles do E. coli exosomes play in microbial physiology?
A: E. coli exosomes serve multiple functions, including:
-
Signaling: They can affect recipient cells' gene expression and help bacteria communicate with one another.
-
Transport: E. coli exosomes transport essential nutrients and enzymes, improving bacterial survival and adaptation in changing environments.
-
Biofilm Formation: Exosomes can contribute to biofilm development by aggregating cells and mediating cell-to-cell interactions.
Q: What potential applications do E. coli-derived exosomes have in biotechnology or environmental science?
A: E. coli-derived exosomes hold significant promise in various fields:
-
Bioremediation: Their ability to encapsulate and transport enzymes could be harnessed for the degradation of environmental pollutants.
-
Synthetic Biology: As natural carriers, they can be used for gene delivery or to engineer microbial communities for industrial applications.
-
Vaccine Development: Their immunogenic properties may be exploited to develop novel delivery systems for vaccines and adjuvants.
Q: How can researchers track or analyze the interactions of E. coli-derived exosomes with other cells?
A: Researchers can use various techniques to track E. coli-derived exosomes and their interactions:
-
Fluorescence Microscopy: Labeling exosomes with fluorescent markers allows visualization of their uptake by target cells.
-
Flow Cytometry: This method can quantify exosome populations and assess their interactions with specific recipient cells.
-
Proteomic and Genomic Assays: Analyzing the cargo of exosomes can help elucidate their role in intercellular signaling and modulation of cellular responses.
Reference
-
Imamiya, Risa, et al. "Escherichia coli-derived outer membrane vesicles relay inflammatory responses to macrophage-derived exosomes." MBio 14.1 (2023): e03051-22. Distributed under Open Access license CC BY 4.0. The image was modified by extracting and using only Part d of the original image and revising the title. https://doi.org/10.1128/mbio.03051-22.
For Research Use Only. Cannot be used by patients.
Related Services:





 Fig.1 Transmission electron microscopy of Escherichia coli-derived exosomes.1
Fig.1 Transmission electron microscopy of Escherichia coli-derived exosomes.1