Haemophilus influenzae-derived Exosome Research & Application
Workflow Research Insights Infrastructure Our Advantages Client Perspectives FAQs
Haemophilus influenzae is a Gram-negative bacterium commonly residing in the human respiratory tract. While generally a commensal microorganism, it has long been recognized for its potential to cause respiratory infections under specific conditions. In recent years, Haemophilus influenzae-derived exosomes (also known as outer membrane vesicles, OMVs) have emerged as a topic of growing scientific interest due to their multifaceted biological roles.
These nanosized vesicles, secreted naturally during bacterial growth, encapsulate lipopolysaccharides, outer membrane proteins, nucleic acids, and other bioactive compounds. They act as communication vehicles, facilitating complex molecular interactions between bacteria and host cells. Their ability to induce immune modulation and to carry antigenic molecules highlights their potential relevance in host–microbe signaling, immune response regulation, and experimental vaccine development.
At Creative Biolabs, our scientific team supports cutting-edge research on Haemophilus influenzae-derived exosomes. Leveraging our expertise in bacterial vesicle isolation and analysis, we help academic and industrial researchers investigate vesicle biology, molecular composition, and functional mechanisms with precision and reproducibility.
Standardized Workflow for Exosome Preparation
At Creative Biolabs, we have established a reliable and flexible workflow for the preparation of Haemophilus influenzae-derived exosomes. Our process integrates stringent quality control and scalable methodologies, ensuring reproducible isolation of vesicles suitable for a variety of downstream research applications.
Standard Workflow (Core Development Process)
-
Culture and Inactivation: Haemophilus influenzae is cultured under optimized aerobic or microaerophilic conditions, followed by heat or chemical inactivation to ensure biosafety.
-
Initial Centrifugation: The bacterial suspension is centrifuged to separate cellular debris from the supernatant.
-
Buffer Exchange: The precipitate is resuspended in a NaCl solution and stabilized in a Tris buffer system to maintain vesicle integrity.
-
Detergent-Assisted Solubilization: A mixture of Tris buffer containing EDTA and sodium deoxycholate is used to extract and liberate exosome-like vesicles from the outer membrane.
-
Ultracentrifugation and Pellet Recovery: High-speed centrifugation is employed to obtain exosome pellets, which are subsequently resuspended in 3% sucrose solution for stabilization.
-
Sterile Filtration: Final preparations undergo filtration to remove contaminants, ensuring the exosome fraction is free of live bacteria.
Optional Add-On Analyses (Subject to Strain Library Availability)
-
Characterization via TEM/NTA – for morphology and size distribution confirmation.
-
Proteomic or Lipidomic Profiling – for comprehensive molecular analysis.
-
Immunogenicity Testing – for exploring host-cell interactions.
Each optional service is tailored according to project scope and species availability within Creative Biolabs' bacterial strain library.
Need tailored exosome isolation or characterization support? Get in touch with us to discuss your experimental goals and available customization options.
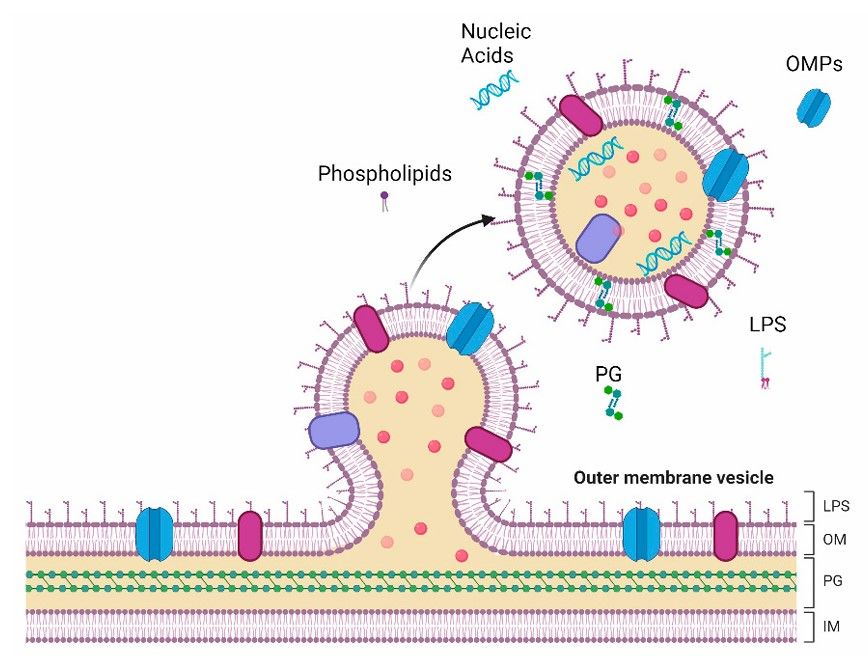 Fig.1 Depiction of EV generation and secretion from the outer membrane of Gram-negative bacteria.1
Fig.1 Depiction of EV generation and secretion from the outer membrane of Gram-negative bacteria.1
Functional Insights: Research on Haemophilus influenzae-Derived Exosomes
Recent studies have provided valuable insight into the structural and immunological behavior of Haemophilus influenzae-derived exosomes. Researchers have identified their significant roles in immune modulation, adjuvant delivery, and bioactive molecular transport. The following table summarizes key research findings reported by scientists studying Haemophilus influenzae-derived vesicles.
|
Research Focus
|
Scientific Findings
|
|
Exosome–Adjuvant Co-delivery Enhances Immune Activation
|
Studies reported that the combination of Haemophilus influenzae-derived exosomes with immunological adjuvants significantly increased antibody titers in animal models compared to adjuvant alone. Specifically, elevated IgG1/IgG2a ratios suggested a bias toward humoral immune stimulation.
|
|
Exosome-Mediated Cytokine Modulation
|
Experiments demonstrated that antigen stimulation following adjuvant-loaded vesicle injection led to enhanced cytokine secretion—particularly IL-4 dominance over IFN-γ and IL-10—indicating activation of a Th2-driven response pathway.
|
|
Intrinsic Antigen Presentation Potential
|
Proteomic analyses revealed that Haemophilus influenzae vesicles carry multiple native bacterial antigens, capable of being recognized by host immune cells, which may explain their immunogenic properties observed in preclinical vaccine studies.
|
|
Controlled Antigen Delivery Systems
|
By encapsulating bioactive molecules within stable vesicular structures, Haemophilus influenzae-derived exosomes demonstrated potential as controlled-release vehicles for experimental drug and antigen delivery applications.
|
|
Comparative Vesicle Composition
|
Comparative characterization of OMVs from different Haemophilus strains revealed notable variation in outer membrane protein profiles, emphasizing the importance of strain selection for reproducible immunological outcomes.
|
These collective findings underline the dual role of Haemophilus influenzae-derived exosomes as both natural mediators of bacterial communication and as promising experimental tools for immunological and microbiological studies. Want to learn how these findings can be applied to your research model? Reach out to our microbial exosome experts for technical consultation.
Research Support Infrastructure at Creative Biolabs
Creative Biolabs has developed a comprehensive research support infrastructure for bacterial exosome studies, ensuring end-to-end technical assistance, from strain cultivation to vesicle isolation, molecular characterization, and data interpretation.
Our platform integrates:
 Customizable Isolation Protocols
Customizable Isolation Protocols
Designed to fit different Gram-negative bacterial species and research goals.
 High-Precision Analytical Tools
High-Precision Analytical Tools
Access to NTA, DLS, TEM, and LC–MS/MS for optional physicochemical and molecular characterization.
 Bioinformatics Support
Bioinformatics Support
Comparative analysis of vesicle proteomes and lipidomes to elucidate biological function.
 Collaborative Data Management
Collaborative Data Management
Secure documentation and data-sharing systems to support transparent project communication.
By providing both core workflows and modular research add-ons, Creative Biolabs ensures flexibility while maintaining scientific rigor across all stages of exosome-related projects. Explore how Creative Biolabs' infrastructure can strengthen your exosome research pipeline, just contact our project specialists today.
Distinct Advantages of Partnering with Creative Biolabs
Collaborating with Creative Biolabs provides tangible benefits for research teams working in microbial vesicle biology.

Scientific Expertise in Gram-Negative Exosomes
Decades of technical experience allow our scientists to optimize workflows specifically for challenging bacterial strains such as Haemophilus influenzae.

Project Customization
Creative Biolabs' modular research structure supports tailored project designs—ranging from small-scale proof-of-concept studies to comprehensive vesicle profiling.

Data Integrity and Reproducibility
Each experiment is conducted under rigorously standardized conditions, with detailed documentation to ensure reproducibility and traceability.

Collaborative Partnership Model
Rather than offering a one-size-fits-all approach, Creative Biolabs emphasizes close scientific dialogue with each client to align goals, methods, and outcomes.

Optional Analytical Expansion
Depending on the available strain database, additional analysis such as proteomics or lipidomics can be integrated into the workflow (optional).
These strengths have made Creative Biolabs a trusted collaborator for academic institutions and research groups pursuing microbiome and exosome-related studies. Interested in forming a long-term partnership? Connect with our research collaboration team to explore custom project solutions.
Client Perspectives and Testimonials
Many research teams that have collaborated with Creative Biolabs have commended our technical reliability, transparent communication, and depth of scientific insight.
"The vesicle preparations were highly consistent between batches, allowing us to reproduce our results effortlessly."
"We truly appreciated the detailed documentation and transparent QC reports - they gave us full confidence in the data integrity."
"The technical support team was exceptionally responsive and knowledgeable, resolving our questions quickly during method setup."
"Collaborating with Creative Biolabs felt like a genuine scientific partnership - their insights helped refine our experimental design and strengthen our conclusions."
Discover how other researchers benefit from Creative Biolabs' expertise - contact us to request sample project summaries or references.
As understanding of Gram-negative bacterial exosomes continues to evolve, Haemophilus influenzae-derived vesicles represent an exciting model for exploring bacterial communication and immunogenicity. Future research is expected to focus on elucidating the molecular mechanisms that govern vesicle biogenesis, conducting comparative functional studies across Haemophilus strains to identify both conserved and strain-specific exosomal markers, and integrating vesicle research with metagenomic analyses to gain deeper insight into microbial ecosystem dynamics. Creative Biolabs remains committed to supporting these advances through methodological innovation, customized research solutions, and continuous scientific collaboration. Join Creative Biolabs in advancing the frontier of bacterial vesicle science - our team is ready to assist your next research milestone.
FAQs
Q: What specific roles do Haemophilus influenzae-derived exosomes play in modulating host immune responses?
A: Haemophilus influenzae-derived exosomes can carry immunomodulatory molecules such as virulence factors, RNA, and proteins that influence host immune responses. They may promote tolerance or immune detection evasion by delivering signals modulating cytokine production or altering antigen presentation. This interaction can lead to a dampened immune response, allowing the bacteria to persist and establish infection.
Q: How do the cargo contents of Haemophilus influenzae exosomes differ from those of other bacterial species?
A: The cargo of Haemophilus influenzae-derived exosomes differs in terms of specific proteins, lipids, and RNA molecules that reflect the unique pathogenic mechanisms and environmental adaptations of H. influenzae. Comparative proteomic studies can reveal distinct virulence factors or regulatory RNAs that may not be present in exosomes from non-pathogenic bacteria, thereby highlighting the tailored strategies H. influenzae employs during infection.
Q: In what ways can the study of Haemophilus influenzae exosomes inform the development of novel diagnostic tools?
A: The study of H. influenzae exosomes can lead to identifying exosomal biomarkers that correlate with infection status or disease progression. By profiling the specific proteins or nucleic acids within these exosomes, researchers can develop non-invasive diagnostic assays that detect H. influenzae infection or monitor treatment effectiveness, significantly improving current diagnostic methods.
Q: What are the implications of Haemophilus influenzae exosomes in biofilm formation, particularly in chronic infections?
A: Haemophilus influenzae exosomes may contribute to biofilm formation by delivering signaling molecules or extracellular matrix components that promote adhesion and aggregation of bacterial cells. This can enhance the persistence of H. influenzae in chronic infections, such as in the respiratory tract, where biofilms are known to shield bacteria from both host immune responses and antibiotic treatment.
Q: How does the biogenesis of Haemophilus influenzae exosomes differ from other bacterial exosomes, and what are the key pathways involved?
A: The biogenesis of Haemophilus influenzae exosomes involves specific membrane budding and fusion processes that may differ from those in other bacteria due to unique outer membrane structures and composition. Key pathways include the involvement of proteins such as bacterial membrane vesicle biogenesis proteins and stress response pathways that modulate exosome release under various environmental conditions.
Q: What potential do Haemophilus influenzae exosomes hold for applications in vaccine development?
A: Haemophilus influenzae exosomes present a promising platform for vaccine development by serving as natural carriers for antigens. Their ability to modulate immune responses and facilitate antigen uptake by dendritic cells could enhance the efficacy of vaccine formulations. Specific exosomal components could also be developed as targeted delivery systems for immunogenic epitopes, potentially inducing a robust and specific immune response.
Q: What challenges are faced in differentiating the functional roles of exosomes derived from pathogenic versus non-pathogenic strains of Haemophilus influenzae?
A: Differentiating the functional roles of exosomes from pathogenic versus non-pathogenic strains can be challenging due to the overlapping characteristics in exosomal composition. Research methodologies must account for variations in environmental conditions and host interactions that may influence exosome production and content. Comparative studies utilizing genomic and transcriptomic analyses, alongside functional assays, are necessary to elucidate these differences effectively.
Reference
-
Magaña, Gisseth et al. "Bacterial Outer Membrane Vesicles: Role in Pathogenesis and Host-Cell Interactions." Antibiotics (Basel, Switzerland) vol. 13,1 32. 28 Dec. 2023. Distributed under Open Access license CC BY 4.0. The image was modified by revising the title. https://doi.org/10.3390/antibiotics13010032
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig.1 Depiction of EV generation and secretion from the outer membrane of Gram-negative bacteria.1
Fig.1 Depiction of EV generation and secretion from the outer membrane of Gram-negative bacteria.1