Custom Protein-PEGylation Conjugation Service
Natural protein therapeutics are often constrained by inherent limitations such as poor solubility in physiological environments, susceptibility to rapid proteolytic degradation, short circulatory half-life due to renal clearance, and unintended immunogenic responses. Protein PEGylation, the covalent attachment of polyethylene glycol (PEG) polymers to protein molecules, has emerged as a powerful technique to address these limitations. This molecular modification transforms fragile proteins into stabilized therapeutics with prolonged bioavailability, enabling reduced dosing frequency and improved clinical outcomes.
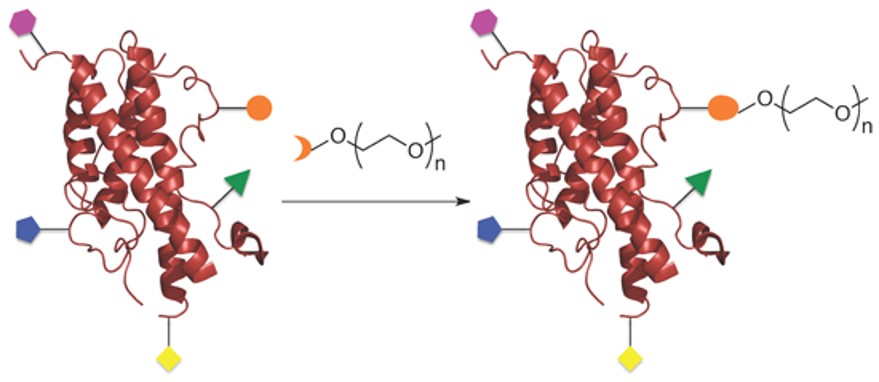 Fig.1 Site-Specific PEGylation of Proteins.1
Fig.1 Site-Specific PEGylation of Proteins.1
This process alters the protein's physicochemical properties through several mechanisms:
- Increased Hydrodynamic Size: PEGylation increases the effective size of the protein, reducing its rate of clearance by the kidneys.
- Reduced Proteolytic Degradation: The PEG chain can sterically hinder the access of proteolytic enzymes, protecting the protein from degradation.
- Masking of Immunogenic Epitopes: PEGylation can shield the protein surface, reducing its recognition by the immune system and lowering immunogenicity.
- Enhanced Solubility: The hydrophilic nature of PEG improves the protein's solubility in aqueous solutions.
Application of Protein PEGylation
Protein PEGylation has found wide applications in various biopharmaceutical fields, including:
- Enhanced Drug Delivery: PEGylation improves the pharmacokinetic and pharmacodynamic properties of protein drugs, leading to increased circulation time, reduced clearance, and enhanced therapeutic efficacy.
- Improved Enzyme Therapy: PEGylation of enzymes can enhance their stability, reduce immunogenicity, and prolong their activity in vivo, making them suitable for enzyme replacement therapy.
- Cancer Therapy: PEGylated proteins are used to deliver cytotoxic agents or modulate immune responses in cancer treatment.
- Diagnostic Applications: PEGylation can be employed to modify proteins used in diagnostic assays, improving their stability and performance.
How Can We Assist Your Project?
Creative Biolabs' Custom Protein-PEGylation Conjugation Service provides tailored solutions to overcome the limitations of protein-based drugs and research tools. We deliver high-quality, PEGylated proteins designed to meet your specific project requirements.
Our Services:
- Wide Selection of PEG Reagents: We provide an extensive range of PEG reagents with various molecular weights and functional groups (including amine, carboxylic acid, NHS ester, thiol, maleimide groups, etc.).
- Customized PEG Reagent Synthesis: Creative Biolabs can synthesize novel PEG reagents with specific properties to meet unique project requirements.
- Targeted Protein PEGylation: We employ site-specific PEGylation strategies to achieve precise modification at desired locations on the protein.
- Validation and Quality Control: We utilize advanced analytical techniques to ensure the identity, purity, activity, and stability of PEGylated proteins.
- Process Scale-up: Creative Biolabs supports the development of robust and scalable PEGylation processes, including process scale-up and preparation of the PEG-modified proteins you need, to provide you with mg to g-scale PEG-modified proteins.
Workflow:
-
Consultation and Project Design
- Analysis of suitable protein conjugation sites
- PEG molecule selection
- Conjugation method selection
- Order Generation
- Conjugate Preparation
- Purification and Characterization
- Product and Certificate of Analysis (CoA) Delivery
Our Technology
- Stochastic PEGylation: We offer random PEGylated protein modifications, primarily targeting the ε-NH2 or α-NH2 groups of lysine residues. Given the high lysine content in most proteins, this approach typically results in a mixture of PEGylated isomers with multiple modified sites. Notably, a significant portion of currently FDA-approved PEGylated drugs are produced using stochastic modification methods.
- Targeted PEGylation: We employ site-specific PEGylation strategies to achieve precise modification at desired locations on the protein. This approach yields a homogeneous product with fewer isomers, better retention of activity, and significantly reduced immunogenicity.
| Modification Type | Description | Key Features/Benefits |
|---|---|---|
| N-terminal amino group targeted modifications | The N-terminal amino group of a protein is often more reactive than lysine ε-amino groups, allowing for selective PEGylation. This can be achieved through pH-controlled reactions or by using PEG reagents specifically designed for N-terminal modification. | Crucial for therapeutic proteins, where modification of other sites might compromise their biological activity. |
| Sulfhydryl group targeted modifications | Cysteine residues contain sulfhydryl (-SH) groups that are highly reactive and can be targeted with thiol-selective PEG reagents, such as maleimides or vinyl sulfones. | Offers a high degree of specificity, as cysteine residues are less abundant in proteins compared to lysine. Particularly useful for proteins with critical lysine residues or engineered cysteine residues. |
| Carboxyl group targeted modifications | Carboxyl groups on aspartic acid or glutamic acid residues can also be used for PEGylation. This often involves activating the carboxyl group with a coupling agent, such as carbodiimide, followed by reaction with a PEG derivative containing a suitable nucleophile. | Can be advantageous for modifying proteins at sites distinct from those targeted by amine or thiol-reactive PEGs, potentially preserving protein function. |
| Enzyme-catalyzed modifications | Enzymes, such as transglutaminases, can be used to catalyze the site-specific attachment of PEG to proteins. For example, transglutaminases can conjugate PEG to glutamine residues. | Occurs under mild, physiological conditions, minimizing the risk of protein denaturation or inactivation. |
Why Choose Us?
- Customized Solutions: We tailor our PEGylation strategies to your specific protein and application, ensuring optimal results.
- Expertise: Our team possesses extensive experience in protein chemistry, PEGylation techniques, and bioconjugation.
- State-of-the-Art Technology: Creative Biolabs utilizes advanced equipment and methodologies to ensure high-quality PEGylated proteins.
- Quality Control: We adhere to stringent quality control standards to guarantee the purity, stability, and activity of our products.
- Comprehensive Support: We provide expert consultation, project management, and ongoing support throughout the project lifecycle.
Customer Reviews
FAQs
1. Q: Will PEGylation affect the biological activity of my protein?
A: Creative Biolabs carefully optimizes the PEGylation process to minimize any potential impact on protein activity. We offer site-specific PEGylation strategies to target non-active sites to ensure the final product meets your requirements.
2. Q: What types of proteins can be PEGylated using your service?
A: Our Custom Protein-PEGylation Conjugation Service applies to a wide range of protein types, including therapeutic, structural, and diagnostic proteins, such as enzymes, antibodies, and cytokines. The suitability of a protein for PEGylation depends on its unique characteristics, and our team will evaluate these to determine the optimal strategy.
3. Q: What is the typical project timeline?
A: Creative Biolabs' typical project timeline depends on project complexity, the protein being modified, and production scale. A general timeline is 4-11 weeks, including: 1-2 weeks for consultation and design; 2-6 weeks for PEGylation and purification; 1-2 weeks for characterization and quality control; and 1 week for final delivery and documentation.
Creative Biolabs provides premium, tailored protein PEGylation services intended to expedite research and development. Inquiries regarding project-specific requirements and further details are welcome.
Reference
- Dozier, Jonathan K., and Mark D. Distefano. "Site-specific PEGylation of therapeutic proteins." International journal of molecular sciences 16.10 (2015): 25831-25864. Under open access license CC BY 4.0, without modification.

