GTOnco™ T Cell Specificity Assay Service
It is well known that the recognition of short peptide sequences presented on MHC class I by the T cell receptors (TCRs) of T lymphocytes is essential for the defenses against malignant cells and pathogens. In adoptive cell transfer (ACT) therapy, the functional activity of large populations of T cells can be redirected against defined targets by antigen-specific TCRs engineering. As a world-renowned service provider for immunotherapy, Creative Biolabs provides high-quality in vitro assays to test the specificity of engineered T cells for interested antigens. GTOnco™ platform also helps our clients design the appreciate project plan to support your gene therapy-based I-O strategies, including CAR-T therapy and TCR modified T cell development.
T Cell Specificity Assay Introduction
T cell specificity is a fundamental mechanism by which the adaptive immune system recognizes and eliminates malignant cells while sparing healthy tissue. This advanced approach allows researchers to decipher the complex mechanisms of T cell recognition, which center on the interaction of the T cell receptor (TCR) with peptide antigens presented by major histocompatibility complex (MHC) molecules on antigen-presenting cells. Each T cell clone possesses a unique TCR capable of recognizing a specific antigenic peptide, and this specificity determines the immune system's ability to distinguish self from non-self, including transformed tumor cells.
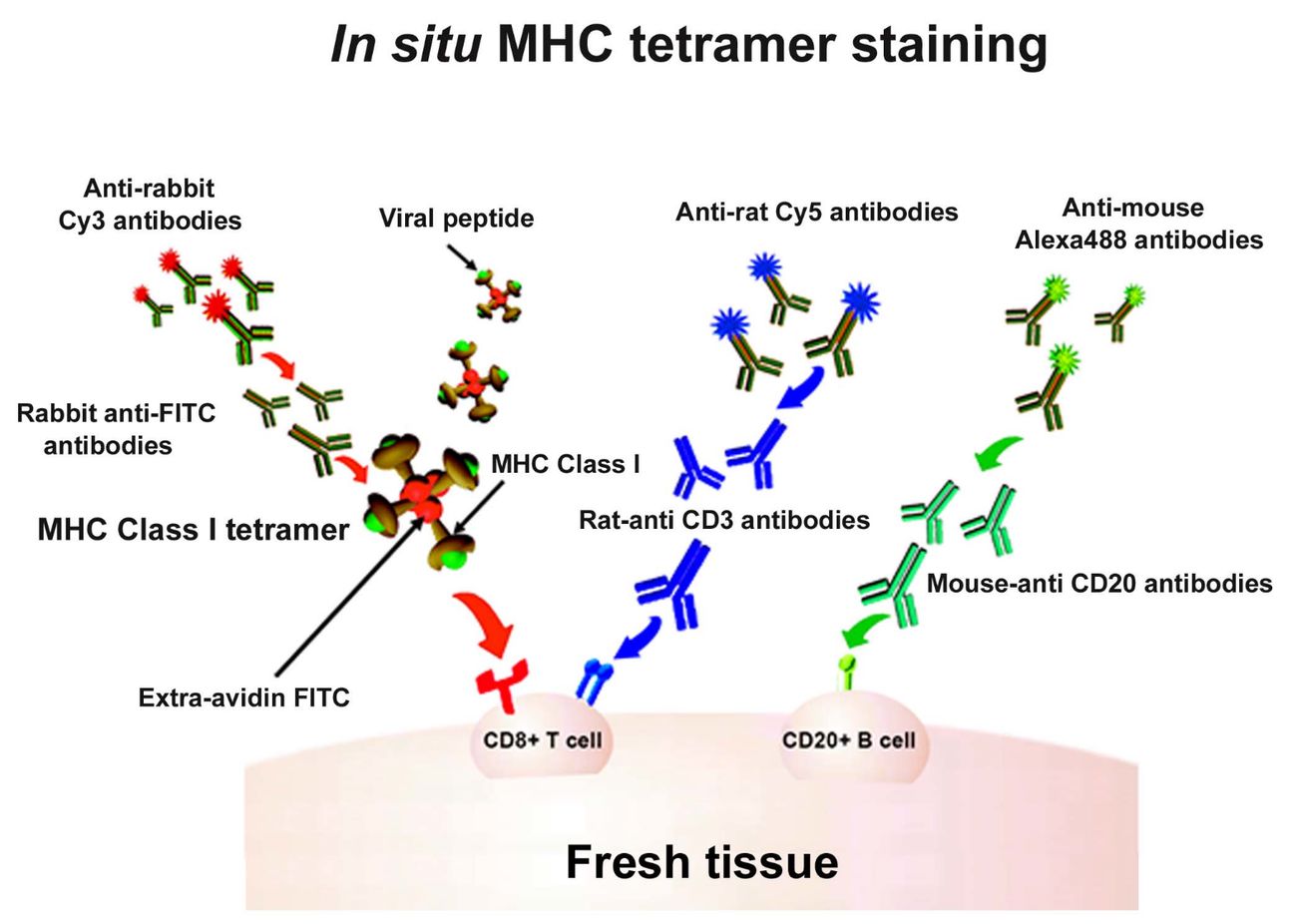 Figure 1 In situ major histocompatibility complex (MHC) class I (MHCI) tetramer staining combined with immunohistochemistry (IHC) to detect virus-specific CD8+ T cells.1
Figure 1 In situ major histocompatibility complex (MHC) class I (MHCI) tetramer staining combined with immunohistochemistry (IHC) to detect virus-specific CD8+ T cells.1
How to Measure Tumor-Specific T Cell Activity Assay
| Functional Readout | Mechanism and Significance | Measurement Technologies |
|---|---|---|
| Cytotoxicity (Killing) | Direct T cell-mediated lysis of tumor cells via release of lytic granules (perforin, granzyme B). This is the ultimate measure of effector function for cytotoxic T lymphocytes (CD8+ T cells). | Cr51 Release Assay (Historical), Flow Cytometry-based Assays (e.g., fluorescent dye labeling of target cells), Live-Cell Imaging Systems, Lactate Dehydrogenase (LDH) Release Assay. |
| Cytokine Secretion | Release of soluble signaling molecules (e.g., IFN-γ, IL-2, TNF-α) upon antigen recognition, driving the immune response. IFN-γ is a key indicator of Th1 and CD8+ effector function. | ELISpot/FluoroSpot (quantifies frequency of secreting cells), ELISA (quantifies bulk cytokine concentration), Intracellular Cytokine Staining (ICS) followed by flow cytometry, Multiplex Immunoassays. |
| Activation Marker Upregulation | Expression of co-stimulatory and activation markers (e.g., CD69, CD25, 4-1BB (CD137)) immediately following TCR engagement, indicating a successful recognition event. | Multi-parameter Flow Cytometry. |
Advanced Technologies of T Cell Specificity Assay
Flow Cytometry-Based Methods
Multiparameter flow cytometry is the leading technique for assessing T cell specificity, capable of simultaneously measuring multiple parameters with single-cell resolution. This broad capability enables complex staining panels that combine pMHC multimer detection with surface phenotyping, intracellular cytokine staining, and viability assessment.
MHC Multimer Technology
The development of pMHC multimer reagents has greatly expanded the scope of T cell specificity profiling. From the initial tetramers to the latest DNA-barcoded multimers, these reagents have progressively increased the number of specificities that can be simultaneously assessed. This approach decouples specificity detection from instrument limitations, potentially enabling analysis of thousands of epitope specificities in small samples (e.g., tumor-infiltrating lymphocytes or limited blood samples).
Enrichment Strategies for Rare Cell Detection
Magnetic-activated cell sorting (MACS) based on activation markers (CD137, CD154) or other surface proteins enables physical enrichment of antigen-responsive cells prior to analysis. This method can increase the detectable frequency of specific cell populations from <1% to >50% of the analyzed sample, significantly improving the resolution of rare cell populations.
Find the Right T Cell Activity Assay for Your Research
Choosing the correct assay depends critically on the therapeutic modality and research question.
| Therapeutic Modality | Primary Research Question | Recommended GTOnco™ Assays |
|---|---|---|
| CAR-T / TCR-T | Functional potency, target specificity, off-target toxicity. | MLTR Assay (Cytotoxicity/Proliferation/Cytokine); TCR Specificity Optimization using Altered Peptide Ligands (APLs). |
| Immune Checkpoint Inhibitors (ICI) | Restoration of exhausted T cell function, reversal of hypo-proliferative state. | Polyclonally Driven CD8 Tex T Cell Assay; Cytokine Response Assay, Cytokine Release Assays (ELISpot/CBA). |
| Cancer Vaccines | Induction of de novo antigen-specific CD4 and CD8 responses, immunogenicity. | CFSE Proliferation Assay; MHC Multimer Staining; ICS for multi-functional T cells. |
| TCR-like Antibodies (TCRm) | Target validation, binding to pMHC complex, delivery efficacy. | T Cell-based High Throughput Multiplexed Assays; T Cell-Mediated Lysis Assay. |
Core Services at Creative Biolabs
Adaptive immune responses are based on the ability of T lymphocytes to respond to specific antigens. Therefore, understanding T cell recognition patterns and T cell specificity in health and disease plays a critical role in the therapeutic efficacy of gene therapy-based I-O drugs development. GTOnco™ platform has developed a variety of in vitro T cell specificity assays to analyze the T-cell epitopes and functions. Such as multi-color flow cytometry, next-generation sequencing and MHC multimer technology for probing antigen specificity is available at GTOnco™. There are three multiplex MHC multimer technologies have been applied in T cell specificity assays, namely heavy-metal-tagged combinatorial encoded MHC multimers, fluorescently labeled combinatorial encoded MHC multimers and DNA-barcode-labelled MHC multimers. These technologies are devised to amplify positive signals and to identify specific TCR specificities.
Quality Isn't an Option—It's Our Guarantee
The GTOnco™ platform adheres to rigorous quality standards to ensure data reliability and reproducibility:
Analytical Validation
Each assay undergoes comprehensive validation, including precision (inter-assay coefficient of variation <15%), accuracy (spike recovery 80-120%), linearity (R² >0.95), and detection/quantification limits.
Reference Standard Implementation
We incorporate well-characterized reference T cell lines and control antigens into each assay, enabling cross-assay standardization and longitudinal monitoring of assay performance.
Multicenter Proficiency Validation
Participation in international proficiency testing programs ensures alignment of our methods with evolving standards for T cell immunity monitoring.
Cross-Platform Validation
Key findings are confirmed using orthogonal methods, ensuring robust conclusions independent of the technology platform.
Our Collaboration Process: A Partnership for Scientific Discovery
At Creative Biolabs, we believe the most impactful scientific discoveries emerge from deep, strategic partnerships. Our collaborative model is designed to serve as an extension of your research team, ensuring our expertise in T cell immunology seamlessly aligns with your unique scientific vision and project goals.
-
Phase 1: Consultation and Experimental Design
Our process begins with an in-depth consultation with your team, led by a dedicated PhD-level project manager and a senior immunologist. Together, we will clarify your primary research question—whether focused on neoepitope validation, monitoring therapy-induced T cell responses, or comprehensive immune profiling.
-
Phase 2: Technical Coordination and Protocol Finalization
With the strategic framework in place, our technical team translates the plan into an actionable protocol. This phase includes:
- Reagent Preparation: Custom production and quality control of pMHC multimers targeting the neoepitope, or configuration of a panel of pre-validated multimers.
- Assay Optimization: Fine-tuning key variables (such as cell stimulation duration, antibody titer, and staining protocol) for your specific sample type.
-
Phase 3: Rigorous Execution and In-Process Quality Control
The wet lab phase of your project will be conducted in our dedicated, controlled laboratory environment. Our commitment to quality during this phase is demonstrated through:
- Standardized SOPs: All procedures adhere to validated and documented methods to ensure reproducibility.
- In-process quality control checks: We monitor key metrics, such as post-thaw cell viability, positive control reactions, and instrument performance, and any deviations are documented and communicated promptly.
-
Phase 4: Comprehensive Data Analysis and Bioinformatics Interpretation
Data generation is just the beginning. Our bioinformatics team transforms complex raw data into biologically meaningful insights.
-
Phase 5: Results Delivery, Reporting, and Strategic Discussion
This final phase ensures the knowledge generated is fully transferable and actionable. You will receive a comprehensive research report with detailed methodology, raw data, publication-ready high-resolution figures and tables, and a clear executive summary.
Client-Centric Service Model
Beyond technical excellence, our service philosophy emphasizes partnership and support:
- Collaborative Approach: We engage as scientific partners, not simply service providers, working closely with clients to refine hypotheses, design appropriate experimental strategies, and interpret complex datasets within their biological context.
- End-to-End Support: We offer comprehensive services, encompassing everything from initial consultation and experimental design to sample processing, data analysis, and result interpretation, providing a seamless client experience.
- Timely Delivery: We understand the importance of timelines in research and therapeutic development, and we have designed a structured process to deliver high-quality data within agreed timelines while ensuring analytical rigor.
Frequently Asked Questions
Q: What sample types are compatible with the GTOnco™ platform?
A: We accept peripheral blood mononuclear cells (PBMCs), isolated T cell subsets, tumor digests, frozen tissue samples, and cultured T cells. Minimum requirements vary by assay, but typically start with 1 to 2 million viable cells for basic analysis. For analysis of rare populations, we recommend using >10 million PBMCs to ensure adequate assay capacity.
Q: Can the platform detect responses to patient-specific neoantigens?
A: Yes, we have extensive experience analyzing responses to personalized neoantigen panels. We require peptide sequences with >70% purity (15-20mers for CD4 responses and 8-12mers for CD8 responses) and can use as little as 100 μg of each peptide.
Q: What controls are included to ensure specificity?
A: Our standard approach includes an unstimulated control (background measurement), a positive control (SEB or anti-CD3/CD28 stimulation), and an antigen-only control (antigen without cells). For multimer-based assays, we include an irrelevant peptide-MHC multimer to assess nonspecific binding.
Q: How is the data delivered and interpreted?
A: We provide comprehensive reports, including raw data files, statistically compared analysis results, and expert interpretation integrated with available literature. Our team offers detailed consultation to interpret the results and recommend next steps.
Connect with Us Anytime!
With professional service and various technology platforms at Creative Biolabs, we provide customized and high-quality T cell specificity assays for our clients all over the world. Our service can be designed to meet your special needs if you have any requirements. If you are interested in our service to support the development of gene therapy-based I-O products, please contact us to learn more information.
Reference
- Abdelaal H M, Cartwright E K, Skinner P J. Detection of antigen-specific T cells using in situ MHC tetramer staining. International journal of molecular sciences, 2019, 20(20): 5165. https://doi.org/10.3390/ijms20205165 (Distributed under Open Access license CC BY 4.0, without modification.)
