Circular RNA (circRNA) Analysis Services
Introduction
Low-abundance RNA, difficulty distinguishing between circRNAs and linear RNAs, and complex functional validation pose challenges in research. Creative Biolabs' Circular RNA Analysis Services address these with advanced expertise in back splicing detection, quantification, and functional validation. We offer comprehensive end-to-end solutions, handling difficult samples, delivering high-resolution, publication-ready data, and going beyond simple detection to accelerate drug discovery and research.
Discover How We Can Help - Request a Consultation
Circular RNA Analysis Services
Circular RNAs (circRNAs) are a class of non-linear RNA molecules that are formed by a backsplicing event, joining a 3' splice acceptor with an upstream 5' splice donor. This unique closed-loop structure makes them highly stable and resistant to exonuclease degradation. Research has revealed that circRNAs are not genetic byproducts but are crucial regulators of gene expression, acting as microRNA sponges, protein scaffolds, and even templates for protein translation. They are increasingly recognized as important biomarkers and therapeutic targets.
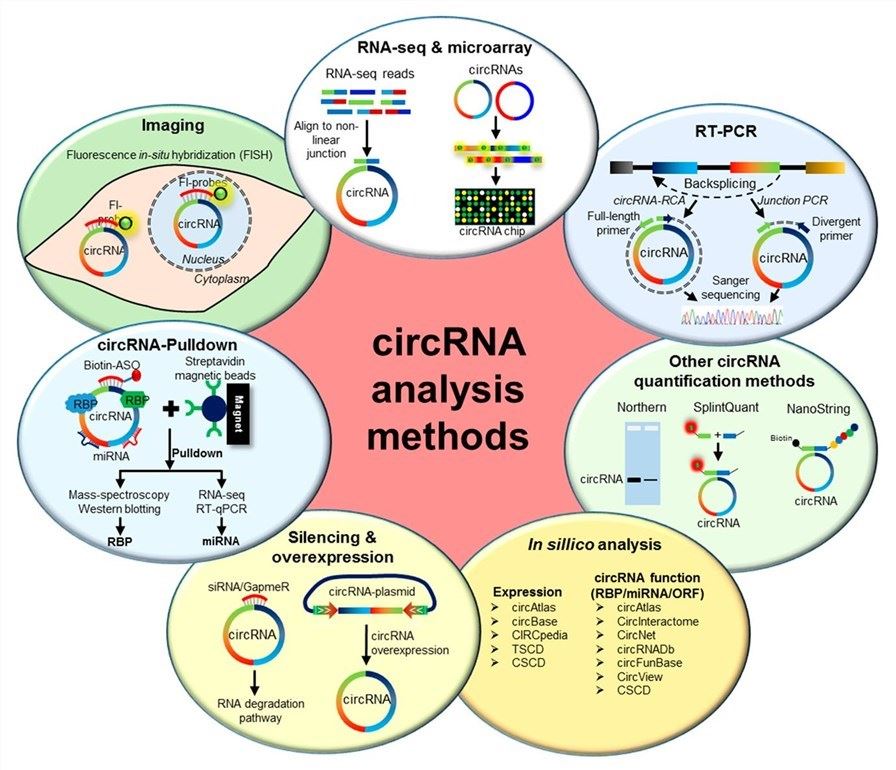 Fig.1 Different circular RNA analysis methods, including RNA sequencing and circular RNA microarray, RT-PCR, Northern blotting, Splint Quant, Nano String, fluorescently labeled probes, etc.1
Fig.1 Different circular RNA analysis methods, including RNA sequencing and circular RNA microarray, RT-PCR, Northern blotting, Splint Quant, Nano String, fluorescently labeled probes, etc.1
In Situ Hybridization Detection for Circular RNAs
Visualize and precisely localize circRNAs in fixed cells/tissues to clarify their cellular function and distribution—nuclear circRNAs may regulate transcription, while cytoplasmic ones might act as miRNA sponges or interact with translation machinery. High-resolution techniques enable single-molecule detail and subcellular mapping, critical for functional studies.
Custom In Vitro Study for Synthetic Circular RNAs
Design and execute tailored experiments to verify synthetic circRNA function, with robust, reproducible data. Assays test biological hypotheses, such as knockdown/overexpression effects on target genes or cell proliferation, and reporter assays to measure miRNA sponge activity, providing mechanistic evidence.
In Vitro Expression Validation for Synthetic Circular RNAs
Confirm successful expression of synthesized circRNAs in various cell lines/models. Using RT-PCR with divergent primers targeting backsplicing junctions, validate correct circularization and stability, laying a foundation for subsequent functional studies.
Workflow
-
Sample Quality Control & Preparation
Check RNA integrity/concentration, remove linear RNA contamination, ensuring high-quality samples for analysis. -
High-Throughput Sequencing & Data Generation
Use advanced platforms to generate deep sequencing data, enabling accurate identification of novel/known circRNAs. -
Advanced Bioinformatics Analysis
Process raw data via specialized pipelines to detect backsplice junctions, quantify circRNAs, and distinguish them from linear RNAs, yielding a comprehensive expressed circRNA list. -
Functional Validation
Validate specific circRNA expression via RT-PCR, northern blotting, etc., confirming sequencing data for publication readiness. -
Comprehensive Data Report
Provide raw/processed data, detailed bioinformatics analysis, and validation results in an understandable, conclusion-supported repo.
Final Deliverables: Upon completion, you will receive:
- A circRNA Expression Profile Report containing identified circRNAs and their expression levels.
- High-quality raw and processed sequencing data for your analysis.
- A Detailed Technical Report summarizing all methods, quality control metrics, and a summary of key findings.
Estimated Timeframe: The typical timeframe for our Circular RNA Analysis Service ranges from 4 to 8 weeks, depending on the number of samples and the complexity of the project. Factors that can influence the duration include sample quality and the level of functional validation required.
What We Can Offer
At Creative Biolabs, we are a leading provider of customized Circular RNA Analysis Services, delivering superior quality and efficiency for your research projects.
One-stop service from initial project design to comprehensive final reports, providing a seamless and efficient workflow.
Specialized workflows designed to handle diverse and complex sample types, including FFPE tissues and low-abundance circRNAs, ensuring reliable data even from challenging sources.
Advanced technical capabilities for both upstream and downstream process development, from precise backsplicing detection to accurate functional validation.
Well-established quality control systems to ensure the integrity of your RNA samples and the accuracy of our data, giving you confidence in your results.
Customized analytical platforms to meet your project's specific needs, whether you require in-depth bioinformatics for novel discovery or targeted validation for a specific circRNA.
High-standard quality control tools are used to precisely quantify and evaluate the quality of circRNA expression profiles.
Guaranteed stability and reproducibility of results through rigorous adherence to best practices in RNA handling and analysis.
Experience the Creative Biolabs Advantage - Get a Quote Today
Customer Reviews

FAQs
How do you differentiate circRNAs from linear RNAs in your analysis?
Our proprietary bioinformatics pipeline is specifically designed to identify the unique backsplice junctions that characterize circular RNAs. This, combined with our linear RNA removal step, ensures we are focusing on and quantifying the true circRNA population in your samples.
Are your services suitable for detecting low-abundance circRNAs?
Yes. We employ high-depth sequencing strategies and specialized enrichment methods that are particularly effective at capturing and quantifying even low-abundance circRNAs, a known challenge in the field.
What is the main advantage of visualizing circRNAs with your In Situ Hybridization service?
Visualization provides critical spatial information that sequencing alone cannot. Our In Situ Hybridization service confirms the presence and subcellular localization of your circRNA, which is essential for determining its function and interaction with other molecules.
How do your services compare to off-the-shelf kits for circRNA analysis?
While kits can be a good starting point, our services provide a complete, end-to-end solution with professional guidance. We manage everything from sample prep to advanced bioinformatics and functional validation, saving you time and ensuring the highest quality results. Our experience allows us to troubleshoot and optimize for complex projects where kits may fail.
Contact Our Team for More Information and to Discuss Your Project
Reference
- Bejugam, Pruthvi Raj, Aniruddha Das, and Amaresh Chandra Panda. "Seeing is believing: visualizing circular RNAs." Non-coding RNA 6.4 (2020): 45. https://doi.org/10.3390/ncrna6040045. Distributed under Open Access license CC BY 4.0, without modification.
