GTOnco™ TCR-pMHC Dissociation Kinetics Assay
To achieve successful immunity, T cells need to be activated through specific interactions between TCRs and antigenic peptides presented by pMHC on antigen-presenting cells. In patients' monitoring or adoptive T cell therapy, the dissociation kinetics of TCRs binding to pMHC have a deep impact on the potency of I-O products. At Creative Biolabs, we are able to use specifically designed reversible fluorescent pMHC multimeric complexes to perform a comprehensive study of TCR-pMHC off-rates. GTOnco™ is pleased to share the advanced technologies and rich experiences for our clients to support their gene therapy-based I-O products development.
TCR-pMHC Dissociation Kinetics Assay Introduction
The interaction between the T cell receptor (TCR) and peptide-MHC (pMHC) is one of the most specific and fundamental recognition events in adaptive immunity. This precise molecular "handshake" determines T cell activation and the initiation of immune responses against pathogens and cancer cells. At the heart of this interaction lies dissociation kinetics—the dynamic rate of dissociation after TCR and pMHC binding. Recent advances in single-molecule biophysics and computational modeling have shown that the dissociation rate (koff) of TCR-pMHC binding is directly related to T cell activation potency, challenging the previous paradigm that primarily focused on binding affinity. Understanding and measuring these kinetic parameters is crucial for developing safer and more effective T cell-based immunotherapies capable of distinguishing healthy and diseased cells with unprecedented precision.
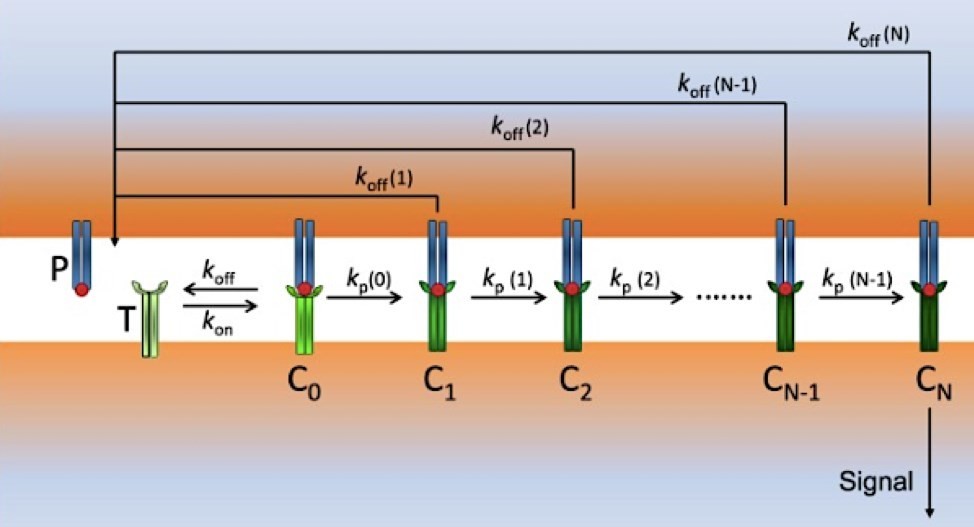 Figure 1. Scheme of modified kinetic proofreading (KPR) model: P (pMHCs) and T (TCRs) bind to form the complex pMHC-TCR (C0) with rate constants kon and koffM/span>.
Figure 1. Scheme of modified kinetic proofreading (KPR) model: P (pMHCs) and T (TCRs) bind to form the complex pMHC-TCR (C0) with rate constants kon and koffM/span>.
TCR-pMHC Complex Overview
The TCR-pMHC complex has emerged as one of the great structural success stories for molecular recognition, allowing for the immune system to interrogate the intracellular proteome via cell surface presentation. The nature of this complex provides important background to understanding the fine details of its dissociation:
Table 1: Key Components of the TCR-pMHC Complex
| Component | Structural Features | Functional Role |
|---|---|---|
| TCR α/β Chains | Variable domains with CDR1, CDR2, CDR3 loops; CDR3 primarily contacts peptide | Antigen recognition and specificity determination |
| CD3 Complex | CD3γ, δ, ε, ζ chains containing immunoreceptor tyrosine-based activation motifs (ITAMs) | Signal transduction and amplification |
| MHC Molecule | Peptide-binding groove with α-helices and β-sheet floor | Peptide presentation and TCR docking platform |
| Bound Peptide | Typically 8-15 amino acids with specific anchor residues | Antigen representation and T cell specificity determinant |
Properties
The TCR-pMHC interface exhibits a relatively small contact area (~1200–2000 Ų) for a protein-protein interaction, but with very high specificity. The small binding interface has implications for the kinetics of binding and dissociation to be rapid, to allow for scanning of many pMHC complexes on the antigen presenting cell.
Mechanosensitive Elements
There are certain components of the TCR-pMHC complex that have been structurally shown to be mechanosensitive. The CD3ε cytoplasmic domain has been shown to dissociate from the membrane in a force-dependent manner, and the pMHC complex itself has been shown to have conformational changes in response to tensile force, which stabilizes the complex by additional hydrogen bonds and hydrophobic interactions.
How Does TCR-pMHC Work?
Features of the TCR-pMHC mechanism that move it beyond a conventional lock-and-key binding interaction include dynamic mechanical parameters to convert molecular binding into cell fate decisions:
- Force-Activated Binding Interface: In contrast to most receptor-ligand pairs, the TCR-pMHC interaction is highly augmented by piconewton-level mechanical forces, typical of the immune synapse.
- Kinetic Proofreading Mechanism: The kinetic correction model suggests that, to ensure productive T cell activation, the lifetime (typically >1-2 seconds) of the TCR-pMHC binding event must be long enough to allow the sequential completion of a series of biochemical steps including CD3 phosphorylation, ZAP-70 recruitment, and LAT phosphorylation.
- Cooperative Mechanical Signaling: Forces mediated through actin cytoskeleton reorganization establish a cooperative mechanical microenvironment which can spatially coordinate the capture bond behavior of multiple TCRs, thus coupling signals across the contact region.
Why Need TCR-pMHC Dissociation Kinetics Assay?
The development of T-cell receptor (TCR)-based therapies has been hampered by unpredictable off-target toxicity and inconsistent efficacy in clinical trials. Traditional binding affinity measurements fail to provide sufficient information to predict T-cell function because they cannot capture the dynamic mechanics characterizing physiological TCR-pMHC interactions. Several key needs necessitate specialized dissociation kinetic analysis methods:
- Predicting T-cell activation potency
- Reducing clinical risk
- Guiding rational TCR engineering design
- Translational biomechanical insights
Services at Creative Biolabs
Our platform has developed high-quality TCR-pMHC dissociation kinetics assay to evaluate possible correlations between T cell function and TCR-pMHC binding kinetics. The approaches based on reversible 2-color multimer have been used to quantify the dissociation rates (i.e., off-rate or koff) directly on living T cells. We provide the custom fluorochrome-conjugated pMHC reagents for our clients to detect the antigen-specific T cells. In addition, to achieve the higher sensitivity, we used pMHC multimer and NTAmer molecules to detect tumor-specific CD8+ T cells which are known to express lower TCR-pMHC affinity/avidity repertoires than virus-specific cells. Our TCR-pMHC dissociation kinetics assay is able to help our clients accurately predict the functional activity of antigen-specific T cells, including their cytotoxicity, proliferation, cytokine release, activating or inhibitory receptor expression, and in vivo anti-tumor potency. Importantly, compared with other frequently used functional assays/metrics, the TCR-pMHC off-rate is a more stable and robust biomarker of T cells potency. At GTOnco™, different parameters of the TCR-pMHC interactions (e.g., KD, koff, kon) will be analyzed to better predict T cell activation and subsequent response potency.
Services offered by Creative Biolabs for TCR-pMHC dissociation kinetics characterization include:
Single-molecule Force Spectroscopy: TCR-pMHC bond lifetime measurement at defined force is determined in the force clamp mode of single-molecule force spectroscopy on BFP and AFM platforms.
Surface Plasmon Resonance (SPR) with Force Modulation: Real-time monitoring of binding kinetics at defined force is achieved on modified SPR instruments equipped with piezoelectric application.
Microfluidic Flow Cell Analysis: Functionality of TCR-pMHC kinetics is measured in microfluidic flow cells that mimic in vivo T-cell scans more closely by imposing hydrodynamic shear.
Structural Dynamics Integration: Services are provided in partnership with structural biology groups for the integration of kinetic parameters with atomic structures.
GTOnco™ Platform Advantages and Highlights
The GTOnco™ Platform by Creative Biolabs represents the convergence of specialized engineering and advanced biophysical analysis, specifically tailored for oncology and T cell therapy.
| Feature | Advantage | Scientific Impact |
|---|---|---|
| Dual-Platform Kinetics | Utilizes both SPR and BLI technologies simultaneously. | Ensures data concordance, eliminates artifact risk, and validates results across orthogonal methods. |
| Low-Concentration Sensitivity | Optimized protocols for accurate detection of sub-nanomolar affinity or high-avidity interactions. | Reliable measurement even for weak physiological interactions and high-affinity engineered TCRs. |
| High-Quality Reagents | Proprietary folding and purification techniques for recombinant TCR and pMHC complexes. | Guarantees $\geq 95\%$ purity and functional integrity, minimizing non-specific binding noise. |
| Custom Peptide Synthesis | Ability to rapidly synthesize and load virtually any peptide sequence into $\mathrm{MHC}$ complexes. | Ideal for neo-antigen discovery and validation of custom therapeutic targets. |
Frequently Asked Questions
Q: What is the relationship between dissociation kinetics and T cell activation potency?
A: Dissociation kinetics, more precisely koff at physiological force conditions, correlates better with T cell activation than binding affinity. Optimal agonists have a binding lifetime between 1-10 s at 5-15 pN force, which allows completion of the TCR-induced cascade reaction. Too short and too long binding lifetimes result in weak activation with a bell-shaped kinetic response curve due to kinetic correction mechanisms.
Q: Can kinetic measurements predict TCR cross-reactivity?
A: Yes. When kinetic data is benchmarked to a panel of related peptides, the probability of cross-reactivity can be predicted with very high accuracy. Weakly discriminative TCRs have a binding lifetime difference of < 3-fold between target and non-target peptides. Discriminative TCRs have 10-100-fold ability to distinguish target from non-target. The principle behind our safety assessment platform is to screen for risky drug candidates at an early stage of the development.
Q: What are the main differences in kinetics between naturally occurring high-affinity TCRs and engineered high-affinity TCRs?
A: Natural TCRs are typically of moderate affinity (KD ~1-100 μM), and their dissociation kinetics is forcing dependent which allows fine grained discrimination between antigens. Many engineered high-affinity TCRs (KD <100 nM) have very low force-sensitivity and establish unusually stable interactions with both target and closely related non-target pMHCs, which is one of the reasons they show higher cross-reactivity. Optimal engineering strategies should focus on preserving/enhancing the kinetic discrimination mechanism of natural TCRs instead of simply trying to maximize affinity.
Customer Review

"The Creative Biolabs team provided impeccable kinetic data for our lead TCR candidate. Their rigorous SPR analysis quickly confirmed that our affinity-mature TCR maintained an ideal binding window—a crucial piece of information that reduced our preclinical risk and accelerated our IND application. The data quality and turnaround time were outstanding."
— Dr. Eleanor Vance, CEO

"We needed a high-throughput solution to screen a yeast display library containing over 300 TCR mutants against a novel MHC class I tumor epitope. Creative Biolabs' BLI platform provided precise k-off values for all candidates in less than three weeks, enabling us to immediately adjust our screening strategy to find the most functional candidates. This was a truly indispensable service."
— Dr. Kenji Tanaka, Lead Scientist
Connect with Us Anytime!
It is known that TCR-pMHC interaction is the keystone of the adaptive immune response. Leveraging our industry expertise, Creative Biolabs is constantly enhancing our in vivo GTOnco™ platform and provides custom and top-quality TCR-pMHC dissociation kinetics assay for our clients to generate new mechanistic insight into disease pathophysiology and improve the process of gene therapy-based I-O drugs development. Contact us today for a proposal to support all of your immuno-oncology research needs.
Reference
- Gálvez J, Galvez J J, Garcia-Penarrubia P. TCR/pMHC interaction: phenotypic model for an unsolved enigma. Frontiers in Immunology, 2016, 7: 467. https://doi.org/10.3389/fimmu.2016.00467 (Distributed under Open Access license CC BY 4.0, without modification.)
