Oncolytic Herpes Simplex Virus Construction Services
Currently, numerous oncolytic viruses, such as herpes simplex virus (HSV), adenovirus, poliovirus, and vaccinia virus are undergoing clinical trials. Nevertheless, HSV-1 stands as the most clinically advanced platform at present. Drawing on years of expertise in immunology and oncology, Creative Biolabs has set up the comprehensive OncoVirapy™ platform and offers oncolytic virus development services. Leveraging OncoVirapy™, our scientists can design, engineer, and efficiently generate oncolytic HSV vectors and viral particles.
Introduction of Herpes Simplex Virus
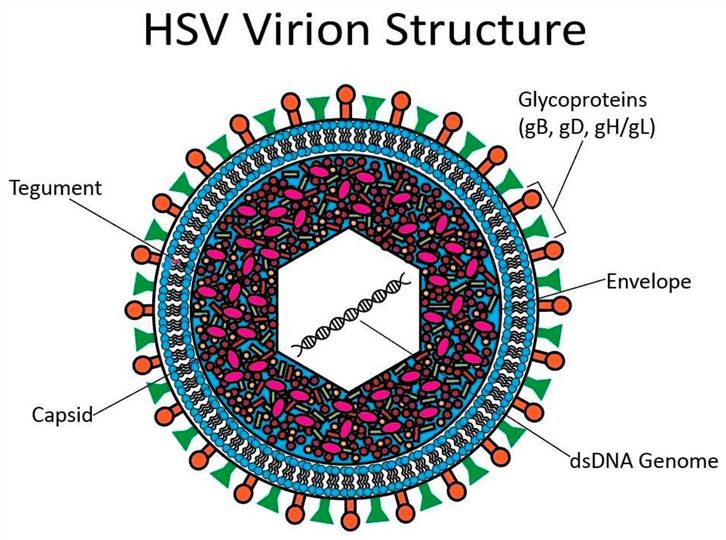
HSV is classified into two distinct serotypes, HSV-1 and HSV-2, primarily based on disparities in their antigenic profiles. The core of the HSV virion consists of a linear dsDNA genome, which is encased within an icosahedral capsid. Beyond the capsid, the HSV virion encompasses two additional structural components: an outer protein tegument and a lipid bilayer embedded with an array of proteins and glycoproteins. HSV gains entry into host cells via either the fusion of its glycoproteins with the host cell membrane or through the process of endocytosis.
Terminal repeats (TRS) at both ends of the HSV genome provide initiation and termination signals for DNA replication and packaging. Genes can be divided into early genes (α, β) and late genes (γ). Early genes are expressed in the early stage of infection and are mainly responsible for virus replication and host regulation. Late genes are expressed late in viral replication and are primarily responsible for the assembly of viral particles.
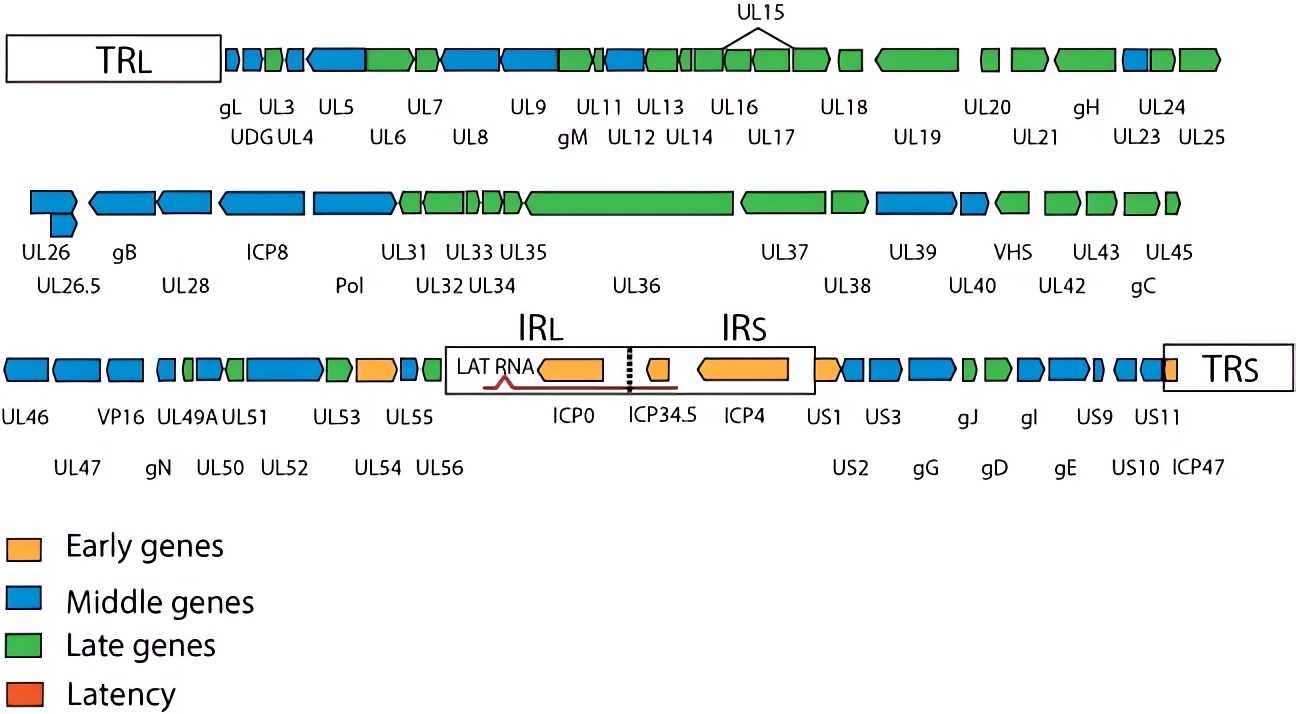 Fig.1 Schematic representation of the genome structure of HSV.Distributed under CC BY 4.0, from SwissBioPics, without modification.
Fig.1 Schematic representation of the genome structure of HSV.Distributed under CC BY 4.0, from SwissBioPics, without modification.
Tab.1 Genome structure, key proteins, and functions of HSV.
| Structure | Unique sequence | UL | Long | TK, ICP6, DNA polymerase |
| Promotes viral DNA replication | ||||
| US | Short | gD, gE, gJ, gL | ||
| Promotes virus binding to host cells | ||||
| Repetitive sequence | RL | Long | Pac sequence | |
| Viral DNA replication and packaging | ||||
| RS | Short | ICP4 | ||
| Promotes viral DNA replication | ||||
| Gene | Immediate-early gene | α | ICP0, ICP4, ICP22, ICP27, ICP47 | Viral gene regulation |
| Early gene | β | ICP6, ICP8 | Viral metabolism | |
| Late gene | γ | Structural proteins of the virus |
Genetic Modifications of Oncolytic Herpes Simplex Virus Vectors
- Genome Editing
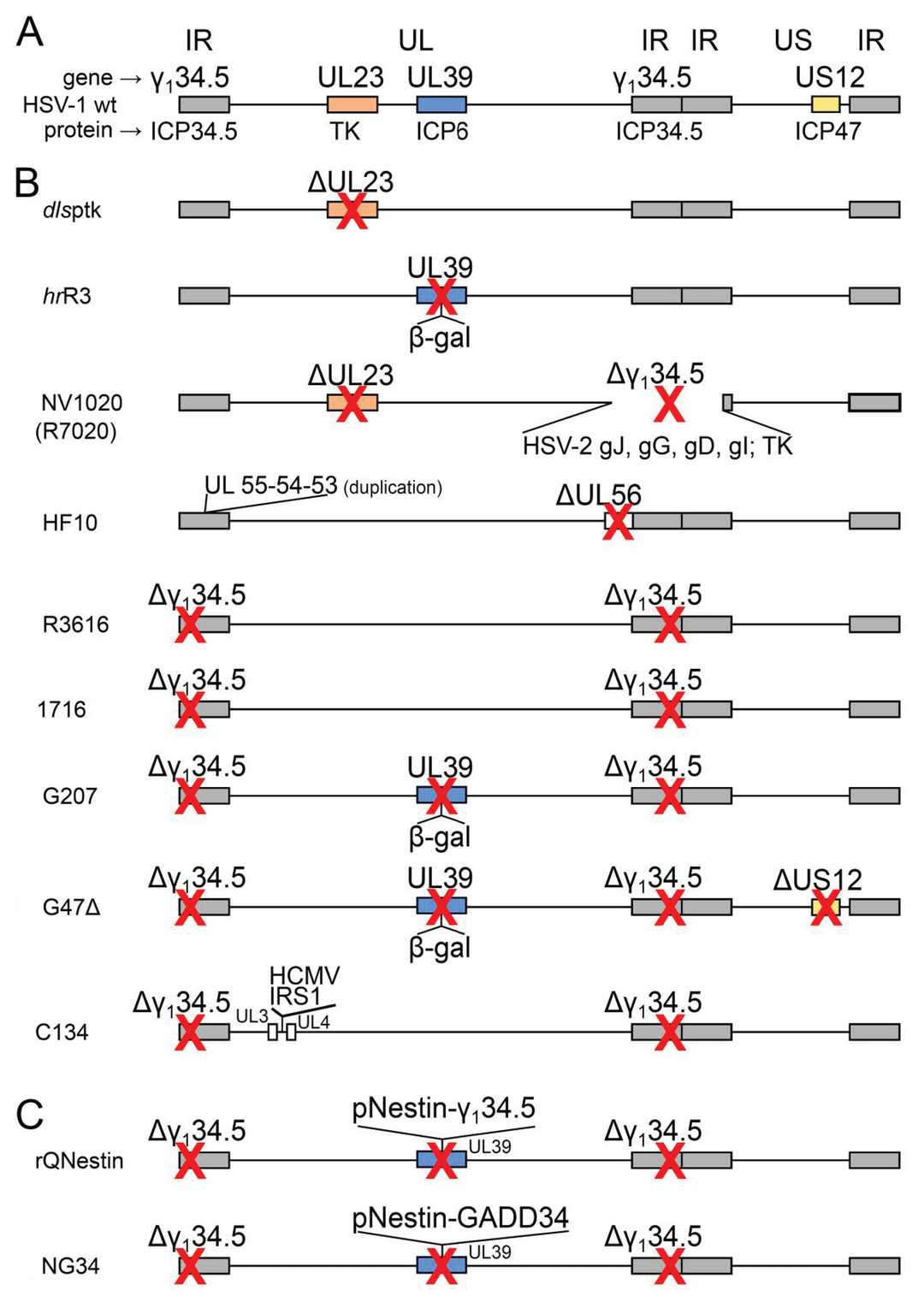 Fig.2 Genomic differences between wild-type and mutant herpes simplex viruses.2,3
Fig.2 Genomic differences between wild-type and mutant herpes simplex viruses.2,3
1. ICP34.5/γ134.5/RL1
The deletion of functional ICP34.5 in HSV-1 can enhance its in vivo safety and mitigate neurovirulence. Nevertheless, this deletion concomitantly leads to a reduction in viral replication. Consequently, engineering the ICP34.5 locus using tumor-specific promoters represents a viable strategy to augment the replication potential of the virus. For instance, promoters of genes preferentially expressed in cancer stem cells (e.g. Nestin and Musashi-1) have been utilized to drive the expression of ICP34.5, thereby facilitating enhanced virus-specific replication within cancer cells.
2. UL23
UL23 encodes the viral thymidine kinase (TK), an enzyme that plays a crucial role in nucleotide metabolism processes essential for viral DNA replication. Proliferating tumor cells characteristically exhibit elevated TK activity. This distinct feature enables TK-deficient HSV to selectively target, infect, and replicate within tumor cells.
3. ICP6
UL39 encodes ICP6, a ribonucleotide reductase that converts ribonucleotides to deoxyribonucleotides, which is essential for HSV DNA replication. Insertion of the lacZ in ICP6 has been used to increase tumor-specific replication of HSV.
4. ICP47
ICP47 down-regulates antigen-presentation activity by inhibiting the transporter associated with antigen processing (TAP) protein. Thus, ICP47 deficiency enables efficient antigen presentation and activates anti-tumor immunity. In oHSV, ICP47 protein mutations enhance immunogenicity and anti-tumor immunity. Moreover, ICP47 deletion promotes early US11 expression, which antagonizes cellular PKR-mediated antiviral signaling, partially compensating for γ134.5 deletion.
- Strategic design of OHSV targeting tumor
Currently, the most pivotal and efficacious arming strategy is accomplished by engineering recombinant genomes. These engineered genomes are designed to express immunomodulatory molecules. Predominantly, cytokines and chemokines (IL-4, IL-12, GM-CSF, etc.) are utilized to arm HSV.
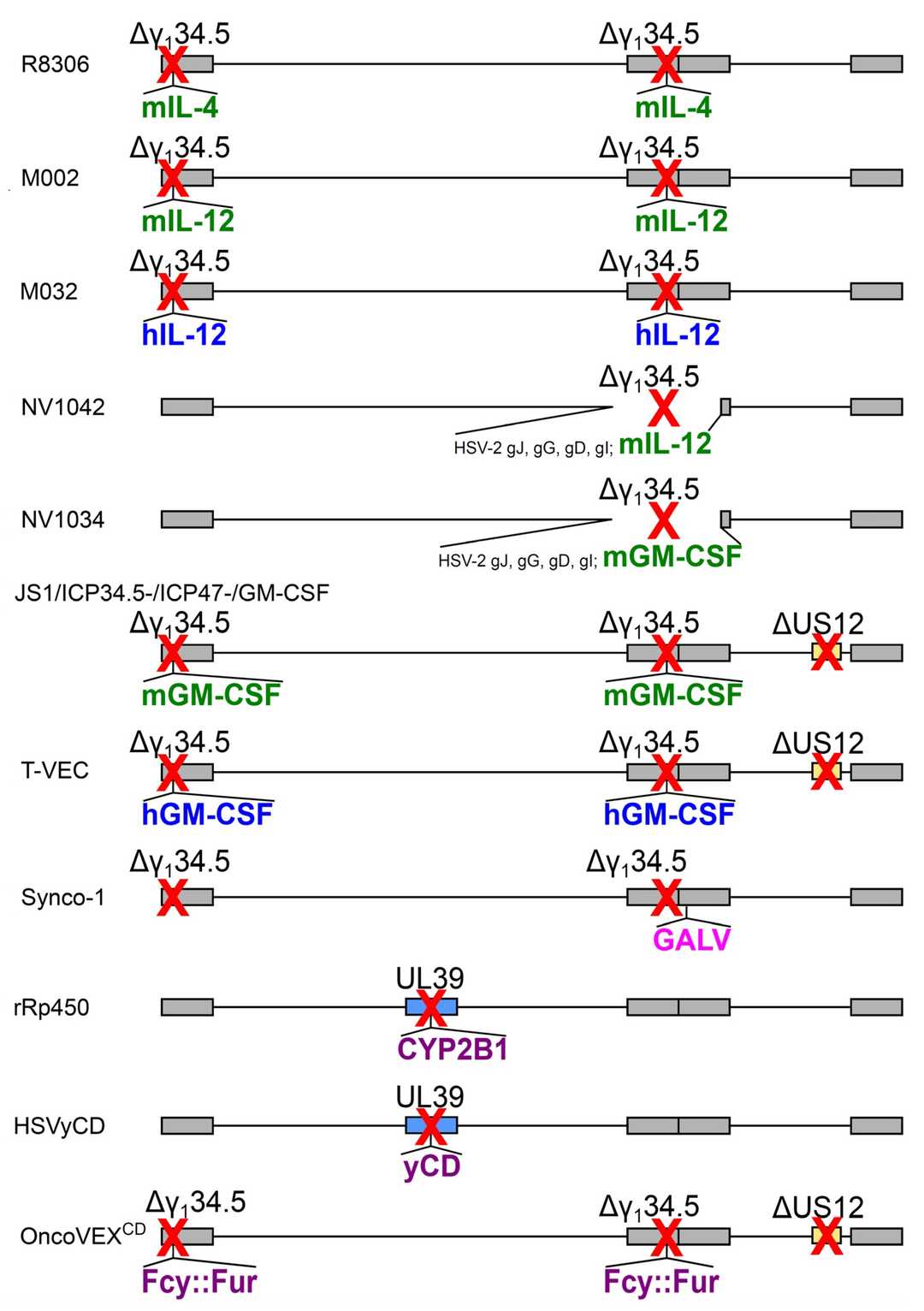 Fig.3 Methods for arming oncolytic herpes simplex virus. 2,3
Fig.3 Methods for arming oncolytic herpes simplex virus. 2,3
Featured Services and Construction Workflow
We provide a comprehensive range of HSV services based on our advanced OncoVirapy™ platform.
- Select the GOI (encoding antibodies, cytokines, chemokines or immune checkpoint inhibitors, etc.) or the genes plan to be deleted for restriction mapping and sequencing verification
- Functional validation of transgenes
- Expression cassette design with the most appropriate promoter to guarantee the high-level expression
- Viral genes engineering (single gene or different genes combination)
- Different construction strategies (homologous recombination, BAC system, etc.)
- HSV amplification, purification, titering services
- HSV whole genome sequencing (WGS) service
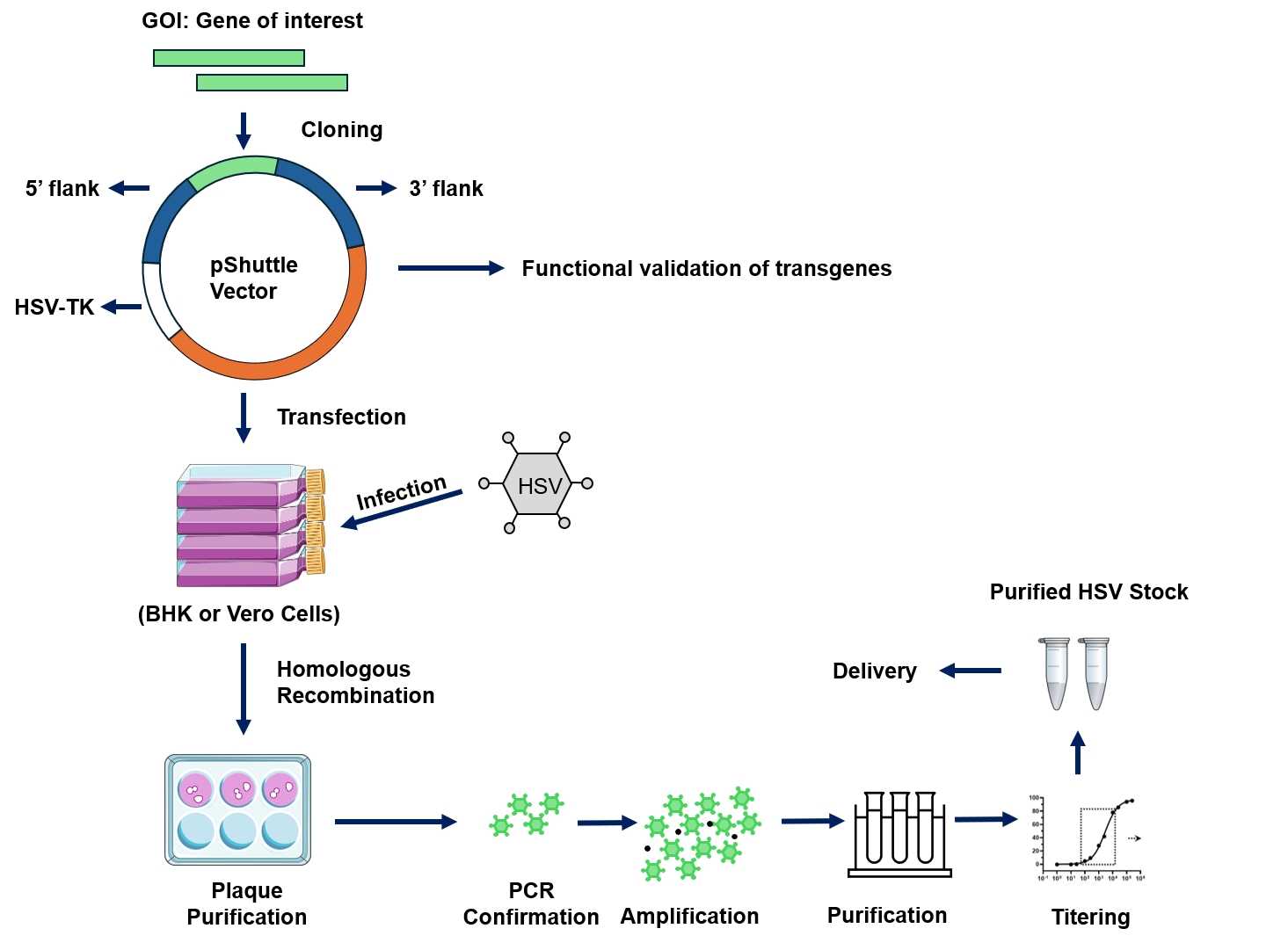 Fig.4 Workflow for Creative Biolabs construction of oncolytic HSV.
Fig.4 Workflow for Creative Biolabs construction of oncolytic HSV.
Cases of Herpes Simplex Virus Genetic Modifications
- T-VEC has been approved for clinical use. Deletion of ICP34.5 and ICP47 in the genome and replacement with GM-CSF attenuated the neurotoxicity of T-VEC and enhanced the replication and expression of GM-CSF in tumor cells.
- G47Δ is a third-generation HSV-1 with triple mutations in γ134.5, ICP6, and α47 genes.
- G207 deleted ICP34.5 in the genome and replaced ICP6 with lacZ, making it replicate specifically in tumor cells lacking P16 tumor suppressor, making remarkable progress in the treatment of brain tumors in children.
- HF-10 naturally lacks the expression of UL43, UL49.5, UL55, UL56, and latency-related transcripts, and overexpresses UL53 and UL54.
- HSV-1716 lacks two copies of the ICP34.5 gene and cannot replicate in terminally differentiated and undifferentiated cells. HSV-1716 can efficiently infect and lye tumor cells.
- VG161 is a novel anti-tumor immune-enhancing oncolytic virus based on HSV-1. It is the first oncolytic virus product carrying four exogenous immune regulatory genes IL12, IL15/15Rα, and PD-L1 blocking peptide (PDL1B) into clinical in the world.
Oncolytic HSV has found extensive application in both pre-clinical research and clinical settings. With the progress of modern genetic engineering techniques, an increasing number of strategies have been uncovered for optimizing the construction of oncolytic HSV. These strategies aim to minimize toxicity while enhancing specificity and efficacy.
Creative Biolabs presents a range of custom oncolytic HSV cloning and construction services, which are characterized by reasonable cost and a rapid turnaround. In addition, we offer in vitro and in vivo validation services for the engineered oncolytic HSV to facilitate the client's whole project.
References
- Sausen, Daniel G., et al. "Herpes Simplex virus, human papillomavirus, and cervical cancer: overview, relationship, and treatment implications." Cancers 15.14 (2023): 3692.
- Menotti, Laura, and Elisa Avitabile. "Herpes simplex virus oncolytic immunovirotherapy: The blossoming branch of multimodal therapy." International journal of molecular sciences 21.21 (2020): 8310.
- Distributed under Open Access license CC BY 4.0, without modification.
