Pathogenicity Manipulation (Attenuation) Service
Introduction
Oncolytic virotherapy, an innovative cancer treatment, has drawn much attention recently as the oncolytic virus can selectively target tumor cells while being harmless to normal ones. But direct live virus administration triggers an immunogenic response, so attenuation of the oncolytic virus is crucial to reduce toxicity to normal cells, lower immune rejection, and enhance tumor cell specificity and replication. The attenuated viruses can synergize with other anticancer therapies like chemotherapy, radiotherapy, or immunotherapy, with multimodal combination therapy emerging as a new trend in cancer treatment.
After years of exploration, Creative Biolabs has established a mature oncolytic virus engineering platform, editing the genome to attenuate pathogenicity, improve drug safety in research and pre-clinical development, and offering full-range pathogenic operations and customized solutions for oncolytic virus development.
Adenoviruses (Ad) with Deletions in E1a/E1b/E3 Gene
Ad is a kind of DNA virus, which has strong infectivity, high safety, easy modification and high titer, etc. Different modification approaches have been developed based on the AD genome structure.
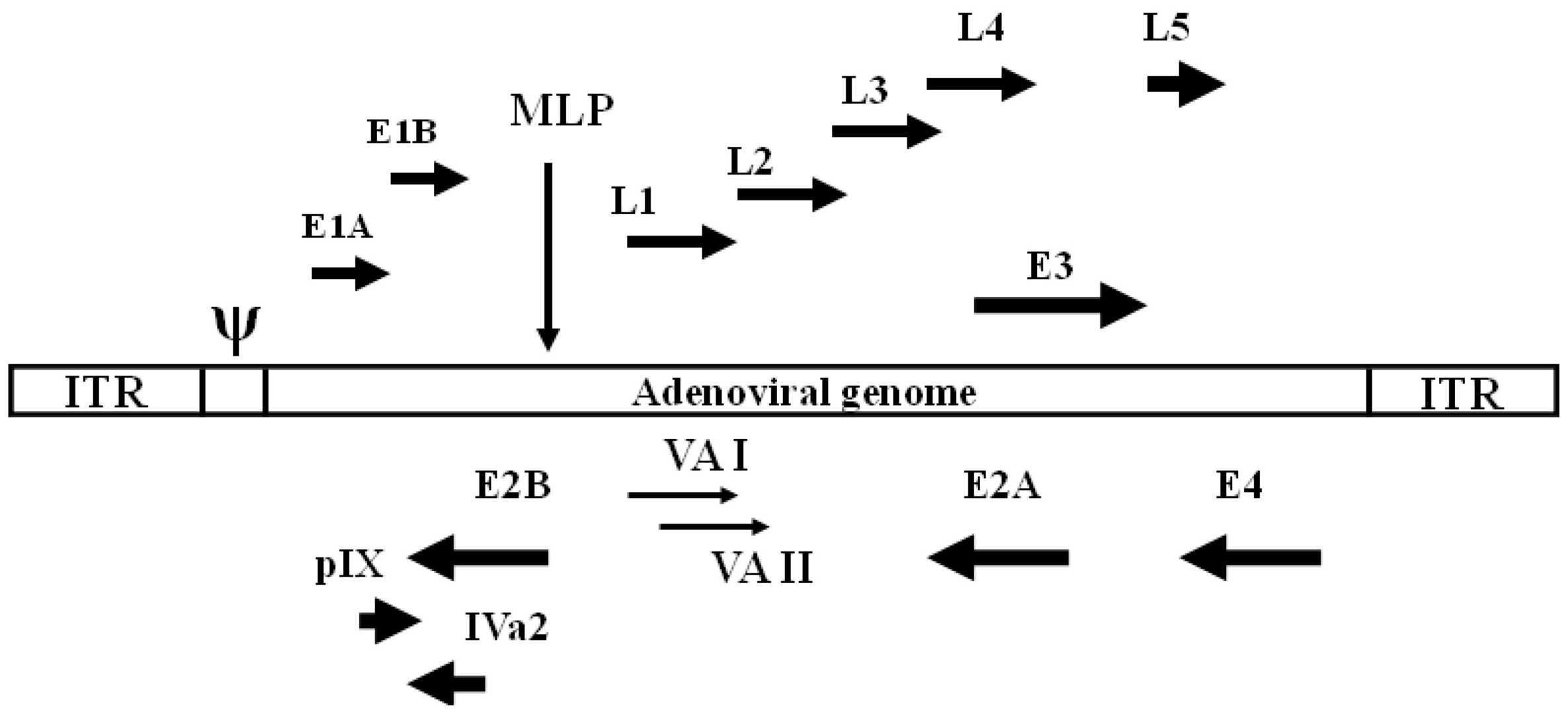 Fig.1 The structure of the adenovirus genome1,4
Fig.1 The structure of the adenovirus genome1,4
-
E1A:
The E1A-CR2 region can bind to the RB gene and provide conditions for virus replication. When the CR2 region of ADs is deleted, the viral E1A in normal cells cannot bind RB to promote viral replication. However, RB regulation is defective in tumor cells. Deletion of CR2 does not affect the replication of ADs in tumor cells, so the specific replication of ADs in tumor cells can be achieved. -
E1B:
E1B contains two proteins, E1B-19K and E1B-55K, which are related to cell cycle regulation. Knockout of E1B-55K restricts adenoviruses from replicating in normal cells. E1B-19k prevents apoptosis by blocking the function of Bax. Simultaneous deletion of 19k and 55k can increase the targeting of the virus and improve the pro-apoptotic ability of the virus. -
E3:
The E3 region contains multiple proteins E3-14.7K, E3-gp19K, ADP (Adenovirus Death Protein), RID, etc.- The gp19 kDa protein inhibits the expression of MHC class I molecules to avoid recognition of infected cells by effector T cells, and E3-14.7K and RID prevent tumor necrosis factor-induced apoptosis. Deletion of E3-14.7K, RID, and E3-gp19K can promote the host immune system to kill infected cells, promote virus replication, and increase the anti-tumor effect.
- ADP can promote cell lysis and mediate virus release. Deletion of this protein slows down the process of virus release, thereby increasing the amount of virus replication in the cell.
Herpes simplex virus (HSV)-1 with Deletions in the γ34.5 and α47 Genes
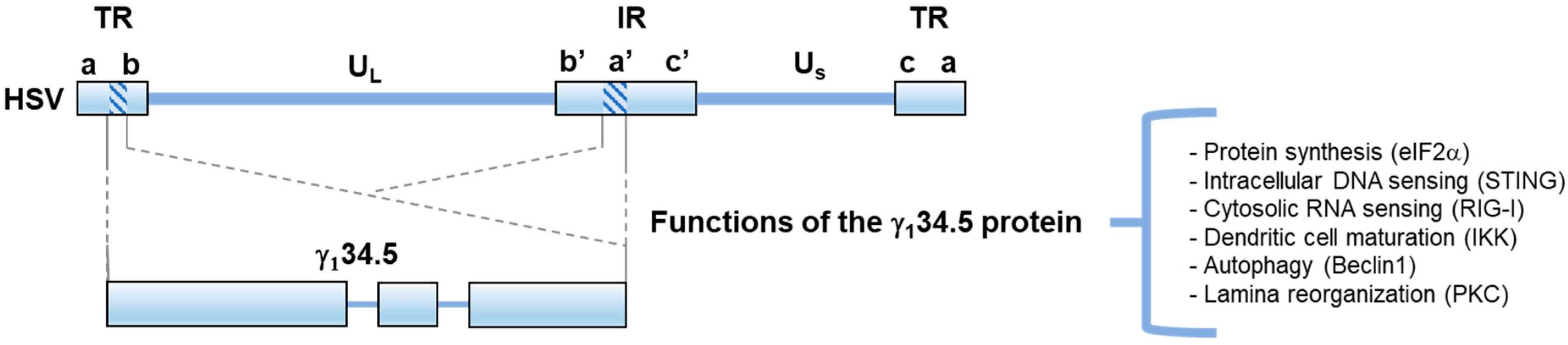 Fig.2 The structure and function of HSV γ134.52,4
Fig.2 The structure and function of HSV γ134.52,4
Specific genes of oHSV-1, such as ICP34.5 (RL1), ICP47, US11, and TK, can be knocked out to enhance the ability of specific growth in tumor cells and viral replication.
-
ICP34.5:
The γ134.5 protein of HSV is multifunctional and can regulate protein synthesis, autophagy, nucleic acid sensing, and virus shedding. Deletion of γ134.5 protein in HSV can improve tumor-specific growth. -
ICP47:
HSV with deletion of the α47 gene shows higher expression of MHC class I molecules than normal HSV. Infected tumor cells are more able to stimulate their matched tumor-infiltrating lymphocytes, which is conducive to tumor elimination. -
ICP6/UL39(E):
ICP6 is a large subunit of ribonucleotide reductase that can be used to synthesize new viral genomes. The mutations in ICP6 can restrict the virus to circulating cells, making it selective for cells. The usual procedure for editing is to inactivate the LacZ fusion protein by inserting it at the C-terminus of ICP6.
Vesicular stomatitis virus (VSV) with M and P mutated
VSV has a relatively small genome size of only 11kb and encodes five proteins: nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and large polymerase protein (L).
 Fig.3 The structure and function of VSV3,4
Fig.3 The structure and function of VSV3,4
- Matrix protein (M):
M is involved in viral assembly, represses host cell gene expression and protects the virus itself from the innate immune response of the host organism. M protein deletion or M51R substitution (VSVΔM51 or VSVM51R) is commonly used.
- Glycoprotein (G):
G is responsible for receptor recognition, cell entry, and viral fusion, making it the primary target immune response that elicits humoral responses. A common modification is the C-terminal truncation of G protein (GΔ28).
Vaccinia Virus (VV) with Mutation in the TK Gene
The pathogenicity of VV can usually be manipulated by mutations in the TK gene. The TK gene encodes thymidine kinase (TK), which not only catalyzes the conversion of deoxythymidine to deoxythymidine but also catalyzes the phosphorylation of some nucleoside analogs. These phosphorylated nucleoside analogs are highly toxic to mammalian cells.
Other Services on Oncolytic Virus Engineering Platform
- Immunogenicity Manipulation
- Antibody-expressing Oncolytic Virus
- Cytokine/Chemokine-expressing Oncolytic Virus
- Immune Checkpoint Inhibitor-expressing Oncolytic Virus
With our well-established oncolytic virus engineering platform, the experienced scientists at Creative Biolabs are dedicated to helping you develop a unique oncolytic virus. We also provide other various services regarding oncolytic virus development. Please feel free to contact us for more information and a detailed quote.
References
- Prusinkiewicz, Martin A., and Joe S. Mymryk. "Metabolic reprogramming of the host cell by human adenovirus infection." Viruses 11.2 (2019): 141.
- Kangas, Chase, Eric Krawczyk, and Bin He. "Oncolytic HSV: underpinnings of tumor susceptibility." Viruses 13.7 (2021): 1408.
- Zinovieva, Margarita, et al. "Oncolytic Vesicular Stomatitis Virus: Optimisation Strategies for Anti-Cancer Therapies." Frontiers in Bioscience-Landmark 29.11 (2024): 374.
- Distributed under Open Access license CC BY 4.0, without modification.
