What Are Comprehensive Aptamer Selection Platforms?
An aptamer selection platform is a systematic workflow, typically based on SELEX, that enriches rare, high-performing DNA/RNA sequences from large combinatorial libraries. The "comprehensive" part matters because different biological questions require different selection environments:
-
Purified protein targets for controlled, quantitative affinity maturation
-
Intact living cells for native conformation, membrane context, and unknown epitope discovery
-
Living animals for physiological distribution, tissue targeting, and in vivo performance pressure
-
Paired binders (two-site recognition) for sandwich assays and higher-confidence detection formats
Great Minds Choose Creative Biolabs
From robust platform design to publication-ready deliverables, our aptamer teams support global innovators with standardized workflows, flexible customization, and consistent data quality.
Partner with Us
Case Studies
Case 1
In vitro selection of an ATP-binding TNA aptamer
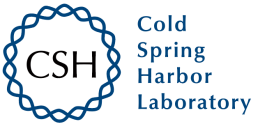
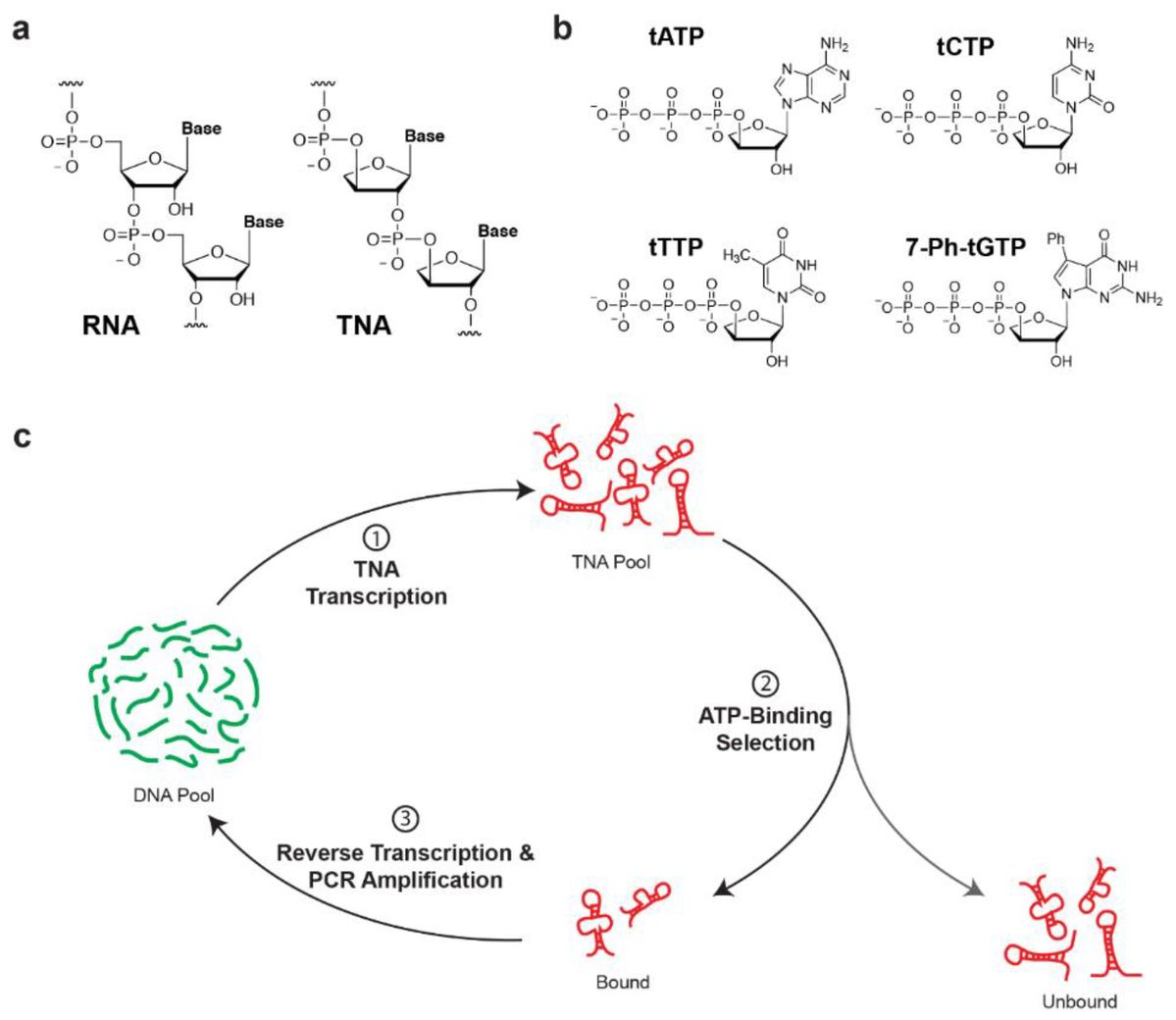
The researchers reported the evolution of an ATP-binding aptamer composed entirely of α-L-threose nucleic acid (TNA). A chemically synthesized version of the best aptamer sequence shows high affinity to ATP and strong specificity against other naturally occurring ribonucleotide triphosphates. Unlike its DNA and RNA counterparts that are susceptible to nuclease digestion, the ATP-binding TNA aptamer exhibits high biological stability against hydrolytic enzymes that rapidly degrade DNA and RNA. Based on these findings, TNA aptamers could find widespread use as molecular recognition elements in diagnostic and therapeutic applications that require high biological stability.
Fig.1 Selection of threose nucleic acid (TNA) aptamers.
1,2
References
-
Zhang, Li, and John C. Chaput. "In vitro selection of an ATP-binding TNA aptamer." Molecules 25.18 (2020): 4194. https://doi.org/10.3390/molecules25184194
-
Distributed under Open Access license CC BY 4.0, without modification.
FAQs
How do you help us choose between protein-based SELEX, whole-cell SELEX, and live animal SELEX?
We start with your intended application (binding reagent, detection, functional blocking, imaging, etc.), the target format feasibility (purified protein availability, membrane presentation, conformational sensitivity), and your real experimental matrix (buffer vs serum-like, cell media, lysate).
Can you develop aptamers that work on native membrane proteins or unknown epitopes?
That’s a core strength of whole-cell SELEX. When epitopes are unknown or the native conformation is hard to reproduce in purified form, cell selection can enrich binders against physiologically presented targets. We’ll help define the most informative control cells and validation plan (e.g., knockdown/overexpression comparisons where available, competition assays, or cross-cell-line specificity profiling).
How do you handle aptamer stability requirements (nuclease resistance, shelf stability, serum tolerance)?
We plan stability from the beginning-either by applying selection pressure closer to your real conditions or by integrating a post-selection optimization pathway. Typical options include chemical modifications (platform-dependent), end-capping strategies, and format recommendations (e.g., labeling choices, immobilization orientation). We also advise on storage and handling to preserve activity across repeated experiments.
What are the typical deliverables?
Deliverables usually include a curated sequence shortlist, enrichment analytics (NGS if used), synthesized aptamers (as requested), and a characterization package aligned with your application—KD/kinetics where appropriate, specificity profiling, and recommended usage conditions.
How do you reduce risk if our target is challenging or our timeline is tight?
We use milestone-based checkpoints (early enrichment signals, interim specificity checks, go/no-go criteria) and can run parallel tracks—e.g., two partition strategies or a protein vs cell approach—when the target is high-risk. This is often more efficient than betting everything on a single selection path and discovering limitations late.
Are your aptamer services suitable for complement-related targets and assays?
Yes. Many complement research programs require binders that perform reliably in complex biological systems and across different assay formats. We can align selection and validation to complement-relevant use cases (e.g., binding to native proteins, cell-surface presentation, or assay pair requirements), while keeping the deliverables focused on research-use performance.