E1A Modification Service
Introduction
At Creative Biolabs, our E1A Modification service addresses key oncolytic adenovirus hurdles: complex engineering, limited tumor specificity, and efficacy bottlenecks. Our OncoVirapy™ Platform develops potent, selective OV vectors that enhance tumor targeting, reduce off-target effects, and boost anti-tumor immunity for safer, effective therapies.
[Discover How We Can Help - Request a Consultation]
E1A Modification
E1A modification is a foundational strategy in the development of tumor-selective oncolytic adenoviruses. The E1A gene of adenovirus is a crucial viral oncogene responsible for initiating the viral replication cycle and driving cellular proliferation. In normal, healthy cells, E1A binds to and inactivates key tumor suppressor proteins like retinoblastoma protein (pRB) and p53, thereby promoting cell cycle progression and creating a permissive environment for viral replication.
However, many cancer cells inherently possess dysfunctional pRB or p53 pathways due to mutations or deletions. By strategically deleting or mutating specific regions within the E1A gene, oncolytic adenoviruses are rendered replication-defective in normal cells, which rely on functional pRB and p53 for cell cycle control. Conversely, in cancer cells where these pathways are already compromised, the viral replication can proceed unhindered, as the necessary cellular machinery is intrinsically dysregulated.
This engineered dependency on cancer cell pathways makes E1A-modified adenoviruses exquisitely tumor-selective. Beyond mere replication control, the E1A region can also be engineered to carry therapeutic transgenes. These "armed" viruses can express a variety of anti-cancer agents upon replication within the tumor, such as:
- Immune-stimulatory molecules: To further enhance the anti-tumor immune response (e.g., GM-CSF, IL-12).
- Prodrug convertases: Enzymes that convert non-toxic prodrugs into cytotoxic agents specifically within the tumor.
- Matrix metalloproteinases or hyaluronidase: To break down the dense extracellular matrix of tumors, improving viral spread and drug penetration.
- Apoptosis-inducing proteins: Such as TRAIL, directly trigger programmed cell death in infected tumor cells.
Therefore, E1A modification transforms a potent viral pathogen into a precision therapeutic tool, capable of delivering multifaceted anti-cancer effects directly to the tumor site, minimizing systemic toxicity, and priming the host immune system for a sustained anti-tumor attack. This strategy is central to developing safer and more effective oncolytic virotherapy.
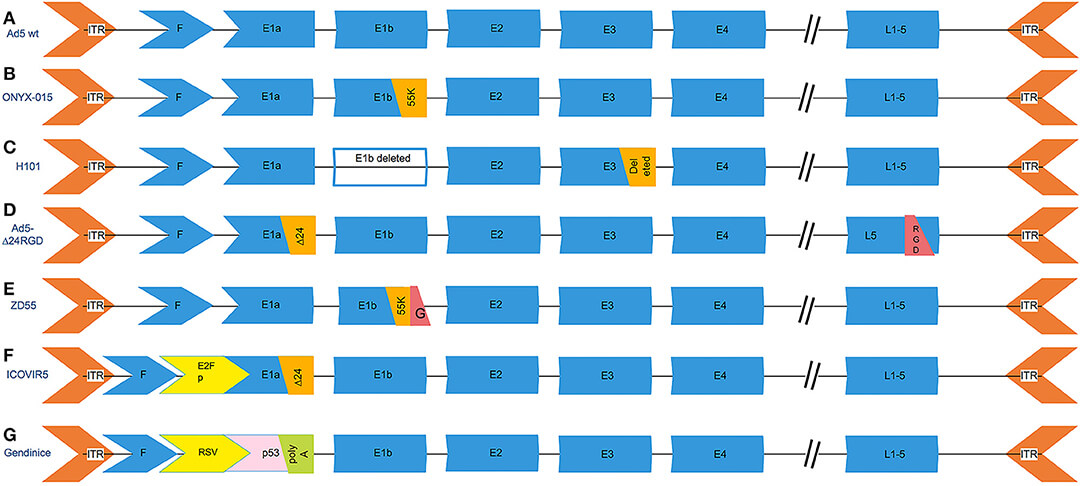 Fig.1 Oncolytic viruses of E1Amodification (D, F, G).1
Fig.1 Oncolytic viruses of E1Amodification (D, F, G).1
Tab.1 The E1A-modified oncolytic adenovirus is currently undergoing preclinical testing.
| Oncolytic Adenovirus | Modification | Efficacy |
|---|---|---|
| AdCV101 | Delete a 24-bp fragment (nucleotides 922–946) in the CR2 region of the E1A gene. | AdCV101 exhibited less cytotoxicity in normal cells than in HCC cell lines. |
| rAd-p53 | E1A gene is replaced with a human wild-type p53 | It can inhibit tumor cell survival signal transduction and specifically kill tumor cells. |
| Ad5-Δ24RGD | A 24-bp deletion (nucleotides 919–943) is introduced in the E1A region. | Replication in cells with a retinoblastoma defective pathway produces a strong antitumor effect in gliomas. |
| ICOVIR-7 | The endogenous E1A promoter has been replaced by the human E2F-1 promoter | Enhanced tumor selectivity, integrin binding, and entry into tumor cells |
| OBP-301 | The E1A promoter was replaced with the human telomerase reverse transcriptase promoter (hTERTp) | Because of the activity of hTERT in tumor cells, oncolytic adenovirus can replicate in large quantities in tumor cells. |
| rAd-sTRII OAd | Deletions of amino acid sequences 4-25 and 111-123 were present in the E1A gene, and sTGFβRII was expressed in the E3B domain. | Highly cytotoxic, sTGFβRⅡ is produced to inhibit TGF-β signaling but does not affect viral replication potential. |
| SG400-E2F/IL15 | The E1A promoter is replaced by E2F-1 promoter, and the coding sequence for interleukin-15 (IL-15) was inserted into the E3 region | Strongly inhibited tumor growth in MDA-MB-231 cells and mice with MDA-MB-231 xenograft tumors. |
| ORCA-010 | The E1ACR2 gene is deleted and the RGD-4C motif is inserted into the fibers. | Enhances integrin binding and has shown efficacy in models of prostate, lung, and ovarian cancer |
Workflow
1. Precise editing of the E1A gene
- Promoter replacement:
Replace the E1A promoter with a tumor-specific promoter (e.g., telomerase hTERT, alpha-fetoprotein) to ensure virus replication only in tumor cells.
- Functional domain deletion:
Deletion of the o CR2 region: disrupts E1A binding to pRb protein and targets tumor cells with aberrant pRb pathways (e.g., 90% of small cell lung cancers).
CR1/CR3 region optimization: regulation of early viral gene expression to enhance tumor selectivity.
2. E1A joint regulatory strategy
- Dual gene regulation: such as E1A and E1B-55K double deletion, dual dependence on p53/pRb deficiency of tumor cells, typical products such as ONCOS-102.
- miRNA response element insertion: The introduction of miRNA binding sites (such as miR-122 binding sites) into the E1A gene inhibits viral replication in normal tissues.
3. Design of E1A enhanced oncolytic adenovirus
- E1A-CR2/RGD double modification:
CR2 depletion enhances tumor selectivity,
Fibrin insertion of RGD peptide enhances infection of αvβ3/5 integrin-positive tumor cells.
- Coexpression of E1A with immunostimulators: For example, E1A coexpresses with GM-CSF to activate the antitumor immune microenvironment.
4. Personalized E1A transformation plan
- Omics data-based customization: Design E1A variants that specifically target mutational pathways based on genomic data from customer-provided tumor cell lines/patient samples.
- High-throughput screening: E1A mutation libraries were constructed, and the best tumor-selective mutants were screened by deep sequencing.
5. Supporting functional verification
- In vitro validation:
Replication selectivity of tumor cells versus normal cells (TCID50 assay),
Cytotoxicity curve (CCK-8/Annexin V assay).
- In vivo validation:
Biodistribution and anti-tumor effect of tumor-bearing mouse model,
Immunohistochemistry was used to analyze the changes in tumor-infiltrating lymphocytes (TILs).
6. CMC grade process development
- Serum-free suspension culture: adapted to HEK293 cell high-density culture (>5×106 cells/mL),
- Double-column purification process: ion exchange and molecular sieve, virus purity > 95% (HPLC detection),
- Lyophilized powder injectable dosage form: storage stability > 12 months at -80°C, clinical grade batch production can reach 1013 VP/batch.
[Experience the Creative Biolabs Advantage - Get a Quote Today]
FAQs
Q: How does E1A modification ensure tumor selectivity?
A: E1A modification exploits pRB/p53 pathway defects in cancer cells. Engineered adenoviruses with E1A alterations are replication-deficient in healthy cells with functional pRB/p53, ensuring efficient replication only in cancer cells with compromised pathways. This achieves tumor-specific targeting and reduces off-target effects.
Q: What types of therapeutic payloads can be incorporated into E1A-modified adenoviruses?
A: Our E1A-modified adenoviruses can be armed with a wide range of therapeutic transgenes. This includes but is not limited to, immune-stimulatory cytokines, prodrug convertase enzymes, proteins that degrade the extracellular matrix, and pro-apoptotic factors.
Q: How does CB ensure the safety and quality of the E1A-modified oncolytic adenoviruses?
A: Our E1A modification strategy enhances tumor selectivity, minimizing replication in healthy tissues. Rigorous quality control ensures high standards in design, production, and purification. All viral vectors undergo thorough characterization for purity, titer, and genetic stability. We provide comprehensive in vitro/in vivo safety assessments.
Q: How does E1A-modified oncolytic virotherapy compare to other cancer immunotherapies?
A: E1A-modified oncolytic virotherapy employs a dual mechanism: direct cancer cell lysis and robust immune activation. Unlike immunotherapies focusing on immune cell modulation, OVs infect and lyse tumors, releasing antigens and danger signals to "activate" the tumor microenvironment's immune response. This synergistic effect is ideal for "cold" tumors and enhances the efficacy of immune checkpoint inhibitors.
Creative Biolabs' E1A Modification service offers a powerful and precise approach to developing next-generation oncolytic adenovirus therapies. By expertly engineering the E1A region, we enable the creation of highly tumor-selective and potent viral vectors capable of direct oncolysis and robust immune system activation. Our commitment to cutting-edge science, rigorous quality, and client-centric solutions positions us as your ideal partner in advancing the future of cancer treatment.
[Contact Our Team for More Information]
Reference
- Abudoureyimu, Mubalake, et al. "Oncolytic adenovirus—a nova for gene-targeted oncolytic viral therapy in HCC." Frontiers in oncology 9 (2019): 1182. DOI: 10.3389/fonc.2019.01182. Distributed under Open Access license CC BY 4.0, without modification.
