Targeted Protein Degradation
What is a Targeted Protein Degrader?
Targeted protein degraders represent a revolutionary class of therapeutic molecules designed to selectively eliminate disease-causing proteins from within cells. Unlike traditional drugs that merely inhibit protein function, these molecules orchestrate the complete removal of their targets. Structurally, a targeted protein degrader is a heterobifunctional molecule, ingeniously composed of three key parts: a ligand that specifically binds to the target protein of interest (POI), another ligand that recruits a cellular E3 ubiquitin ligase, and a chemical linker that covalently connects these two functional ends. This unique architecture enables these degraders to act as molecular bridges, bringing a specific protein and a component of the natural protein disposal system of cells into proximity to initiate targeted degradation.
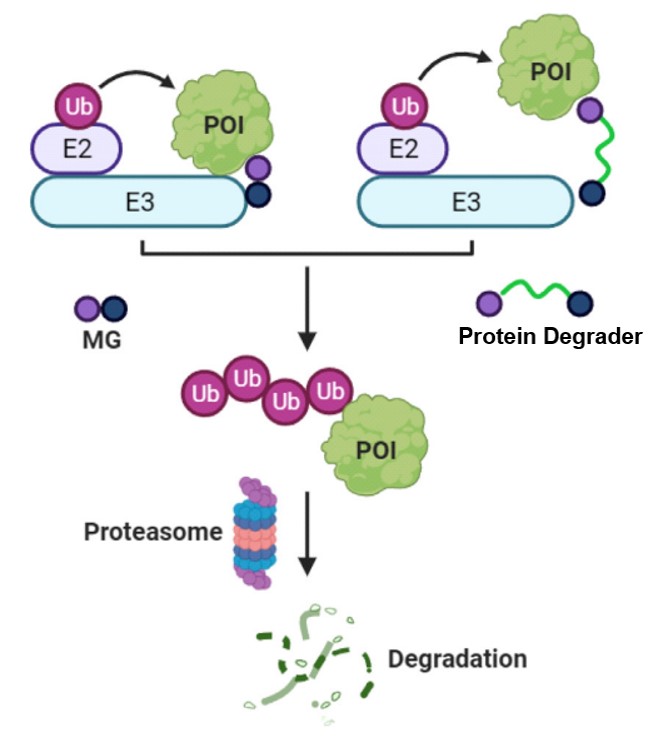 Fig.1 Mechanism of degrading undruggable targets by protein degraders.1
Fig.1 Mechanism of degrading undruggable targets by protein degraders.1
The Genesis and Evolution of Protein Degrader
The concept of using such molecules for therapeutic benefit was first introduced in 2001 by Crews, Deshaies, and colleagues, who envisioned hijacking the cell's ubiquitin-proteasome system (UPS). Early iterations, often referred to as peptide-based degraders, utilized peptide sequences to recognize either the POI or the E3 ligase. While these pioneering molecules validated the degradation concept, their development was often hampered by poor cell permeability and stability. A significant advancement came with the development of all-small-molecule protein degraders, which replaced peptide components with more drug-like small organic molecules, enhancing their cellular uptake and in vivo applicability. This transition marked a pivotal moment, paving the way for the first successful in vivo demonstrations of targeted protein degradation and, eventually, the entry of protein degraders into human clinical trials, heralding a new era in drug discovery.
The Design of Protein Degraders
The design of an effective protein degrader primarily considers the following aspects:
- Target Selection
Protein degraders, leveraging their event-driven mechanism, offer unique target opportunities. Ideal targets often include traditionally "undruggable" proteins like STAT3, drug-resistant mutants such as the BTK C481S mutation, and proteins with amplified gene expression or overexpression. Moreover, these molecules exhibit the capability to selectively degrade specific protein isoforms even with high binding site homology among family members, a significant advantage over traditional inhibitors. They can also effectively target scaffolding proteins that are difficult to inhibit directly. Additionally, protein degraders are being explored to address protein aggregates implicated in various neurodegenerative diseases. This broadens the scope of treatable diseases beyond those addressable by traditional small molecule inhibitors.
- E3 Ligase Selection
Crafting effective protein degraders requires E3 ligands with adequate affinity to recruit active E3 ligases without disrupting ubiquitination. This is challenging due to limited ligand availability for the vast majority of the 600+ human E3 ligases. Cereblon (CRBN) and Von Hippel-Lindau (VHL) are prevalent choices because their small molecule binders are accessible, suitable for linker conjugation, enable flexible degradation of diverse targets, and their widespread expression supports systemic protein degradation. IAP and MDM2 are other commonly engaged E3 ligases. A significant focus is now on developing ligands for tissue- or tumor-specific E3 ligases to mitigate potential systemic toxicity and enhance protein degrader tolerability by confining protein degradation to the intended therapeutic site.
- POI Ligand Design
The selection of POI ligands in protein degrader design critically determines degradation efficiency and selectivity. An ideal ligand must fulfill two key criteria: 1) preferential binding to solvent-exposed regions of the target protein (e.g., allosteric pockets or non-catalytic domains) to provide spatially compatible linker attachment sites, thereby avoiding steric clashes that could impede productive ternary complex formation; 2) presence of modifiable chemical groups (e.g., amines, carboxyls) to enable linker conjugation without compromising binding affinity.
- Linker Design
The linker's type and length, typically varying alkyl or polyethylene glycol (PEG) chains, are crucial for optimal ternary complex formation and subsequent degradation. Extremes in linker length can hinder complex assembly or reduce degradation efficacy. Beyond merely connecting, linkers can interact with protein surfaces, and conformational constraints within the linker can enhance degrader activity by reducing flexibility or locking in a bioactive state, as seen in degraders for SMARCA2/4. Furthermore, the linker structure can be engineered to modulate the pharmacokinetic properties of the protein degrader.
Mechanism of Action and Distinct Advantages of Protein Degraders
Protein degraders operate through an elegant mechanism that co-opts the cell's endogenous protein quality control machinery. Upon entering a cell, a protein degrader molecule simultaneously binds to its target POI and an E3 ubiquitin ligase, forming a transient ternary complex. This induced proximity facilitates the transfer of ubiquitin molecules from an E2-conjugating enzyme (associated with the E3 ligase) onto lysine residues of the POI. The polyubiquitinated POI is then recognized by the 26S proteasome, which unfolds and degrades it into small peptides, effectively eliminating it from the cell. A key advantage is the degrader's catalytic mode of action: once the POI is degraded, the protein degrader is released and can engage another POI molecule, allowing for sustained degradation at sub-stoichiometric concentrations. This "event-driven" pharmacology contrasts with the "occupancy-driven" mechanism of traditional inhibitors. Furthermore, protein degraders offer the potential to target proteins previously deemed "undruggable" (e.g., those lacking enzymatic active sites, like scaffolding proteins or transcription factors), overcome inhibitor resistance mechanisms (such as target mutation or overexpression), and achieve improved selectivity due to the specific protein-protein interactions required for ternary complex formation.
Creative Biolabs possesses extensive experience in bioconjugation and related chemical biology techniques. We are well-equipped to provide comprehensive protein degrader development services, from initial design and synthesis to preclinical evaluation, helping our partners accelerate their innovative therapies.
Reference
- Zhang, Chao et al. "Targeting the undruggables-the power of protein degraders." Science bulletin vol. 69,11 (2024): 1776-1797. doi:10.1016/j.scib.2024.03.056. Distributed under Open Access license CC BY 4.0. The image was modified by extracting and using only part of the original image.
Related Services
- Custom Small Molecule Synthesis and Modification Service
- Custom Protein-Small Molecule Conjugation Service
- Oligonucleotides-Small Molecule Conjugation Service
- Custom Small Molecule-Particle Conjugation Service
