The complement system is made up of a large number of distinct plasma proteins including enzymes, receptors, and complement inhibitors. Most of the proteins are normally inactive, but in response to the recognition of molecular components of microorganisms, they become sequentially activated in an enzyme cascade, the activation of one protein enzymatically cleaves and activates the next protein in the cascade.
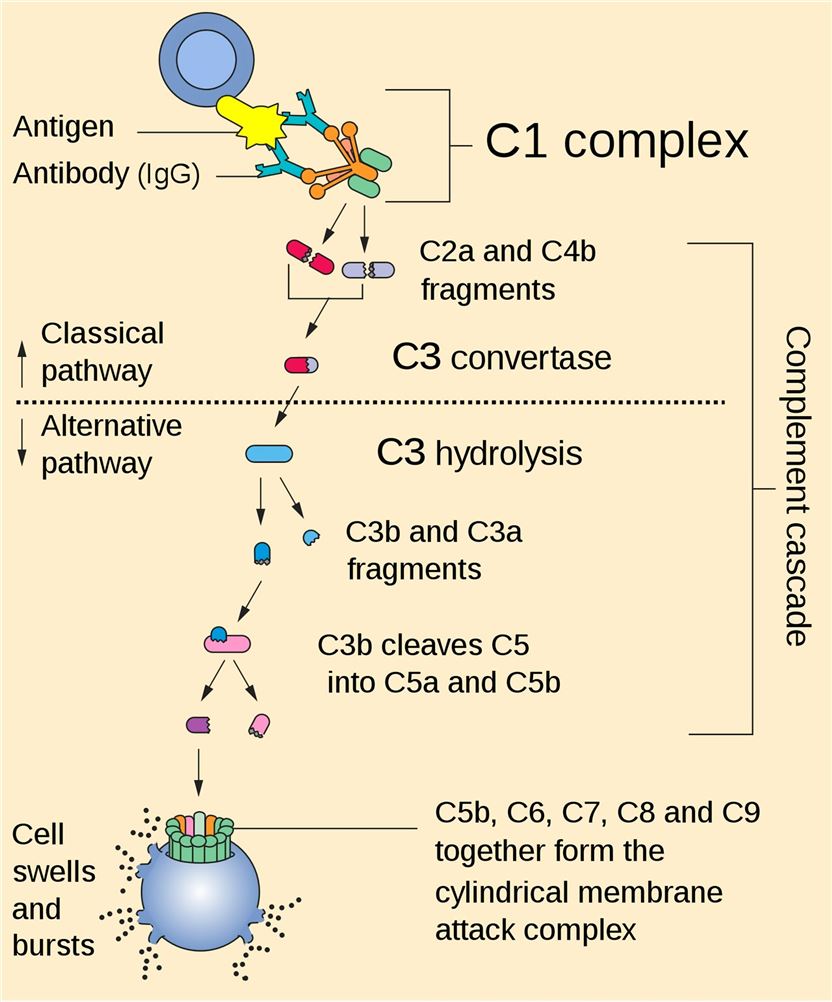
Fig. 1 Complete cascade.1
-
Complement Component Protein
Complement proteins are soluble and circulate in the blood in an inactive form. In response to an activating mechanism, some complement proteins are transformed into proteases and cleave other specific complement members, initiating amplification cascades. Complement can be activated through three different pathways, classical pathway, lectin pathway, and the alternative pathway. All three pathways converge at C3, resulting in the formation of the C3a and the C3b fragment. Factor I cleaves C3b into iC3b and C3f. The conformational rearrangement of iC3b generates binding surfaces for the interaction with complement receptors CR2, CR3, and CR4. The C3b fragment can also bind to other complement peptides to form the C5 convertase. The resulting cleavage of C5 is the beginning of the terminal pathway of the complement: C5 is cleaved into C5a and C5b. The latter forms a complex with C6 and C7, which then binds with C8. The combination of the C5b678 complex with 1 to 18 C9 molecules, termed membrane attack complex (MAC). Creative Biolabs offers comprehensive set of complement test services for global clients.
-
Complement Regulator of Complement System
The powerful effector functions of complement have the potential to harm the host. The activation of classical pathway and lectin pathway can cause autologous injury under some certain situations. For example, deposition of C3b via the alternative pathway activation and amplification is nondiscriminatory and, if not properly regulated, can damage host cells rapidly. Human beings have developed variety regulatory proteins to regulate the location and activity of complement.
The complement regulatory proteins include membrane-bound complement regulatory proteins and soluble complement regulatory proteins. The majority of complement regulatory proteins consist of individually folded short consensus repeats (SCRs) containing about 60 amino acids per SCR including 4 conserved cysteine residues that form intradomain bonds. The former mainly includes membrane cofactor protein (MCP), complement receptor 1 (CR1), decay-accelerating factor (DAF), complement receptor of the immunoglobulin superfamily (CRIg), thrombomodulin and CD59. The latter includes complement factor I (CFI), complement factor H (CFH), C1 inhibitor (C1-INH), C4 binding protein (C4BP), vitronectin, carboxypeptidase N, clusterin and so forth.
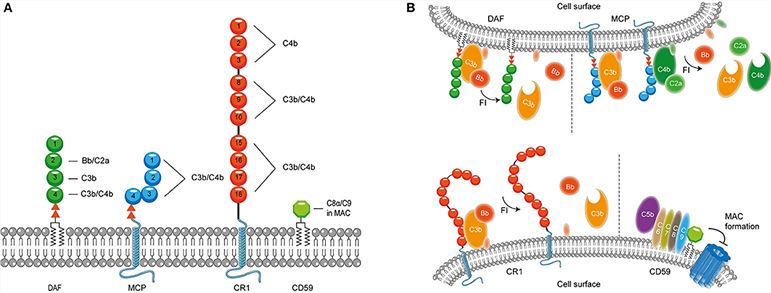
Fig. 2 Membrane bound complement regulatory proteins.2, 4
Complement Deficiencies
Complement deficiency is an immunodeficiency of absent or suboptimal functioning of one of the complement proteins, leading to overwhelming infection and sepsis. Complement deficiencies are inherited and most of them are autosomal recessive traits with the exception of a deficiency of the C1 inhibitor (autosomal dominant) and properdin deficiency (X-linked). Diseases of an autoimmune nature are frequently associated with a deficiency of one of the early complement components C1, C2, or C4 and CR1 receptors. Deficiency of the early complement components would prevent the activation of C3 and the fixation of a resulting C3 cleavage product. Under this circumstance, erythrocytes would be unable to participate in the transport of the immune complex to the reticuloendothelial system.
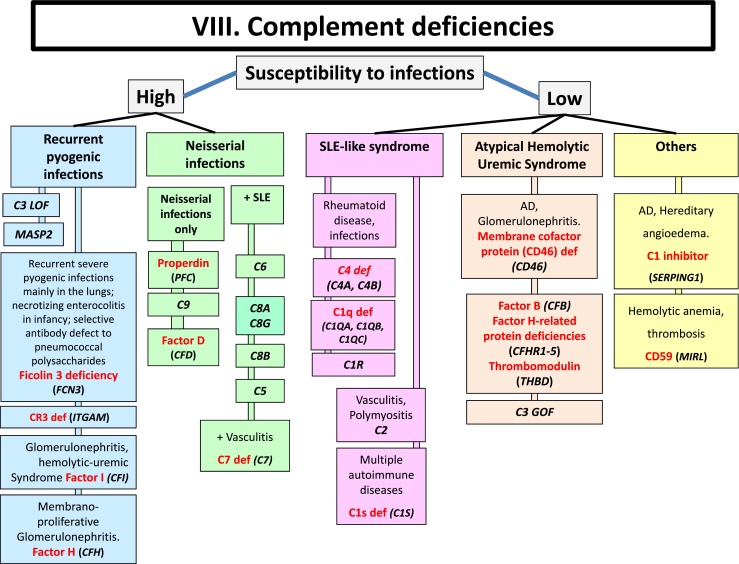
Fig. 3 Complement deficiencies.3, 4
Creative Biolabs offers a number of complement system-realted services and complement system-realted products. If additional help is needed, please directly contact us and consult our technical supports for more details.
References
-
From Wikipedia: https://commons.wikimedia.org/wiki/File:Complement_pathway.svg
-
Hovingh, Elise S., Bryan Van den Broek, and Ilse Jongerius. "Hijacking complement regulatory proteins for bacterial immune evasion." Frontiers in Microbiology 7 (2016): 2004.
-
Bousfiha, Aziz, et al. "The 2015 IUIS phenotypic classification for primary immunodeficiencies." Journal of clinical immunology 35 (2015): 727-738.
-
under Open Access license CC BY 4.0, without modification
Related Product
For Research Use Only.
Related Sections:



