The field of immunology has undergone a paradigm shift with the advent of immunotherapy and personalized medicine. T-cell receptor (TCR) clonality assessment has emerged as a cornerstone technology for decoding adaptive immune responses, tracking disease trajectories, and evaluating therapeutic efficacy. However, researchers face significant challenges in capturing the full spectrum of TCR repertoire diversity, detecting minimal residual disease with high sensitivity, and translating complex genomic data into clinically actionable insights. At Creative Biolabs, we bridge this gap with our cutting-edge TCR Clonality Assessment Service, leveraging over 20 years of biological expertise to deliver precise, reliable, and interpretable data that drives innovation in oncology, autoimmunity, and infectious disease research.
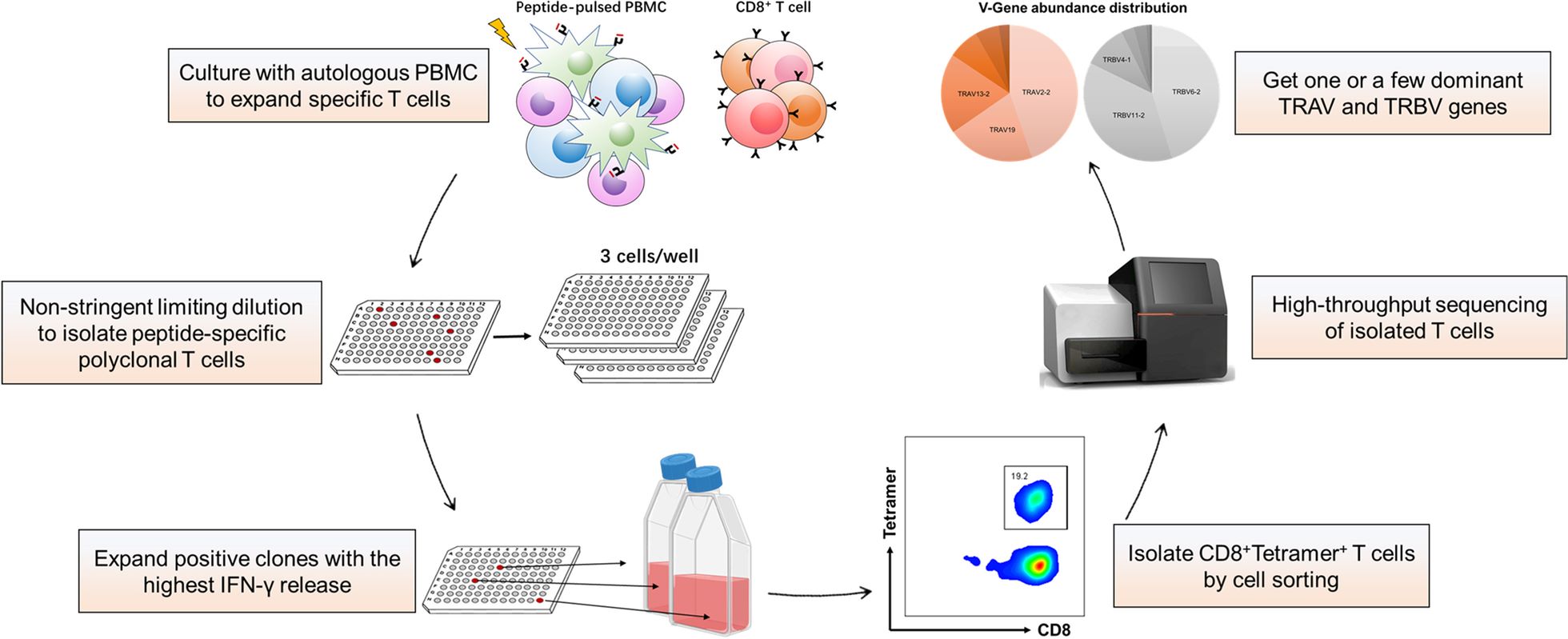 Fig.1 Schematic of the nonstringent TCR cloning method1,2
Fig.1 Schematic of the nonstringent TCR cloning method1,2
At Creative Biolabs, we offer comprehensive TCR clonality analysis solutions built upon a multidimensional cutting-edge technology platform, specifically designed for your high-end immunology research. From precise protocol design to deep data insights, our team remains your trusted research partner, delivering reliable, scalable, and highly interpretable data to accelerate your entire immunotherapy and disease mechanism research projects.
Selecting the right partner for TCR clonality assessment is pivotal to the success of your projects. Creative Biolabs stands out through its unwavering commitment to excellence, backed by over 20 years of industry leadership.
Our team of PhD-level biologists and bioinformaticians brings deep knowledge in immunology and genomics, ensuring scientifically sound interpretations and recommendations
With optimized wet-lab protocols and computational algorithms, we achieve detection limits as low as 0.001% for rare clonotypes, minimizing false positives and negatives
Leveraging automated workflows, we handle projects of any size—from pilot studies to large cohorts—without compromising on quality or timelines
From project design to data delivery, our customer success team provides personalized guidance, helping you maximize the value of your investment.
We have streamlined our service process to ensure efficiency, transparency, and client satisfaction. Below is an overview of the key steps involved:
We begin by understanding your objectives, sample types, and experimental design. Our experts collaborate with you to tailor the service to your specific needs
Clients submit samples (e.g., blood, tissue, or FFPE blocks), which undergo rigorous QC checks for DNA/RNA integrity and concentration
Using optimized kits, we construct sequencing libraries and perform high-coverage NGS on Illumina or similar platforms
You receive a detailed report via a secure portal, followed by a debriefing session to discuss findings and potential next steps
Our TCR clonality assessment service is not merely a collection of processes, but is built upon an integrated platform that deeply integrates cutting-edge wet lab techniques, proprietary bioinformatics algorithms, and big data insights. This ensures every data point you receive carries the depth and reliability of our technology.
![]() Patented Primer Design & Multiplex PCR System
Patented Primer Design & Multiplex PCR System
![]() Dedicated Bioinformatics Cloud Platform
Dedicated Bioinformatics Cloud Platform
We understand raw data alone does not yield insights. Therefore, every client gains access to our proprietary bioinformatics cloud platform:
Interactive Visualization: Generate professional charts—including clonal distribution maps, dynamic evolutionary trajectories, and diversity index comparisons—via drag-and-drop operations without programming knowledge.
![]() Internal TCR Sequence Reference Database
Internal TCR Sequence Reference Database
Leveraging years of experience processing hundreds of thousands of samples, we have built and continuously optimized an extensive internal TCR sequence reference database.
Our TCR Clonality Assessment Service offers a comprehensive suite of solutions tailored to meet the diverse needs of academic institutions, pharmaceutical companies, and clinical laboratories. Each component is meticulously crafted to ensure accuracy and scalability.

High-Throughput TCR Sequencing
Utilizing next-generation sequencing (NGS) platforms, we deliver deep profiling of TCR beta (TCRB) and TCR alpha (TCRA) chains. This includes full-length sequencing to capture complete CDR3 regions, enabling detailed clonotype identification and quantification.

Advanced Bioinformatics Analysis
Our proprietary pipelines perform robust data processing, including error correction, clonotype assembly, and diversity metrics calculation. We also offer comparative analysis across samples to track clonal dynamics over time or between conditions.

Customized Reporting and Visualization
Clients receive intuitive reports featuring clonotype frequency distributions, Shannon diversity indices, and phylogenetic trees. Interactive dashboards allow for easy exploration of data, facilitating hypothesis generation and decision-making.

Integration with Multi-Omics Data
For a holistic view, we can correlate TCR clonality data with transcriptomic, genomic, or proteomic datasets, providing insights into immune microenvironment interactions.
TCR clonality assessment has broad applicability across multiple domains, enabling breakthroughs in research and clinical practice:
|
Monitor T-cell responses to checkpoint inhibitors, CAR-T therapies, or vaccines; identify predictive biomarkers for treatment efficacy. |
|
Characterize aberrant T-cell expansions in conditions like rheumatoid arthritis or multiple sclerosis to uncover pathogenic mechanisms. |
|
Track antigen-specific T-cell repertoires during infections (e.g., HIV, COVID-19) or vaccination campaigns. |
|
Assess immune reconstitution and detect graft-versus-host disease-related clonal expansions in hematopoietic stem cell transplant recipients. |
|
Support preclinical and clinical trials by evaluating immunogenicity and safety profiles of novel therapeutics |
| Feature | Standard Service | Premium Service |
|---|---|---|
| Sequencing Depth | 50,000 reads/sample | 500,000+ reads/sample |
| Clonotype Detection Limit | 0.1% | 0.001% |
| Analysis Complexity | Basic metrics | Advanced statistics & multi-omics integration |
| Turnaround Time | 3-4 weeks | 2-3 weeks |
| Expert Consultation | Standard report | Dedicated scientist support |
Hear from researchers and industry leaders who have leveraged our service to achieve their goals:
What sample types are supported?
We accept peripheral blood mononuclear cells (PBMCs), fresh or frozen tissue, FFPE blocks, and extracted DNA/RNA. All sample types undergo rigorous nucleic acid quality and concentration validation to ensure data reliability.
What is the core technological principle behind this service?
Our service is centered on next-generation sequencing (NGS) technology. By designing multiplex PCR primers targeting conserved regions of the T cell receptor (TCR) gene, we specifically amplify gene fragments containing the Complementary Determining Region 3 (CDR3), which determines T cell specificity. CDR3 represents the most variable region within the TCR, functioning as a "molecular barcode" where its unique sequence defines a specific clonal type. By performing high-throughput sequencing of these CDR3 sequences and conducting bioinformatics quantitative analysis, we can precisely map the composition and dynamics of the entire T cell population within your sample.
How can we perform integrated analysis with multi-omics data?
The data formats we provide are compatible with mainstream transcriptomic and genomic analysis workflows. For example, you can combine TCR clonality data with RNA-seq data from the same sample to analyze the correlation between specific T cell clonality and immune checkpoint gene expression, cytokine profiles, and other factors within the tumor microenvironment. This enables a deeper, systems-level understanding of immune response mechanisms.
How do we distinguish meaningful clonal amplification from background noise in data analysis?
We employ a rigorous bioinformatics workflow: First, we remove amplification and sequencing errors; Second, we only recognize clonal types as genuine if they exceed a threshold percentage (e.g., 0.001%) of total sequencing reads. Finally, by aligning against experimental background controls or healthy donor data, we further filter out common sequencing background or non-specific amplification products, ensuring reported clonal amplifications possess biological significance.
Q: How do you ensure data reproducibility?
Our protocols include spike-in controls, duplicate sequencing, and cross-validation with orthogonal methods. We adhere to strict QC standards throughout the workflow.
Q: Can you handle longitudinal study designs?
Yes, we specialize in time-series analyses and paired sample comparisons, providing insights into clonal evolution and persistence.
Q: What bioinformatics approaches do you employ?
We use established tools (MiXCR, IMGT) complemented by custom algorithms for clonotype calling, diversity analysis, and visualization.
Q: How is data confidentiality maintained?
We implement encrypted storage, secure transfer protocols, and strict confidentiality agreements compliant with GDPR and HIPAA regulations.
References
For any technical issues or product/service related questions, please leave your information below. Our team will contact you soon.
All products and services are For Research Use Only and CANNOT be used in the treatment or diagnosis of disease.
 NEWSLETTER
NEWSLETTER
The latest newsletter to introduce the latest breaking information, our site updates, field and other scientific news, important events, and insights from industry leaders
LEARN MORE NEWSLETTER NEW SOLUTION
NEW SOLUTION
CellRapeutics™ In Vivo Cell Engineering: One-stop in vivo T/B/NK cell and macrophage engineering services covering vectors construction to function verification.
LEARN MORE SOLUTION NOVEL TECHNOLOGY
NOVEL TECHNOLOGY
Silence™ CAR-T Cell: A novel platform to enhance CAR-T cell immunotherapy by combining RNAi technology to suppress genes that may impede CAR functionality.
LEARN MORE NOVEL TECHNOLOGY NEW SOLUTION
NEW SOLUTION
Canine CAR-T Therapy Development: From early target discovery, CAR design and construction, cell culture, and transfection, to in vitro and in vivo function validation.
LEARN MORE SOLUTION