Ectodermal cells derived from induced pluripotent stem cells (iPSCs) serve as a critical starting point for generating various specialized cell types such as neurons, astrocytes, oligodendrocytes, melanocytes, and keratinocytes. These derivatives are essential for neurobiology, dermatology, developmental biology, and high-throughput drug screening applications. At Creative Biolabs, we provide end-to-end solutions for ectoderm differentiation, delivering highly defined, reproducible, and scalable protocols that meet the growing demand for human-origin research models.
The ectoderm is one of the three primary germ layers formed during embryogenesis. It gives rise to the central and peripheral nervous systems, the epidermis, and associated appendages. Directed differentiation of iPSCs into ectodermal lineages involves the inhibition of mesodermal and endodermal signals, mainly through dual-SMAD inhibition, which suppresses both BMP and TGF-β signaling pathways. This process mimics the natural embryonic development environment, enabling high-efficiency ectodermal fate specification.
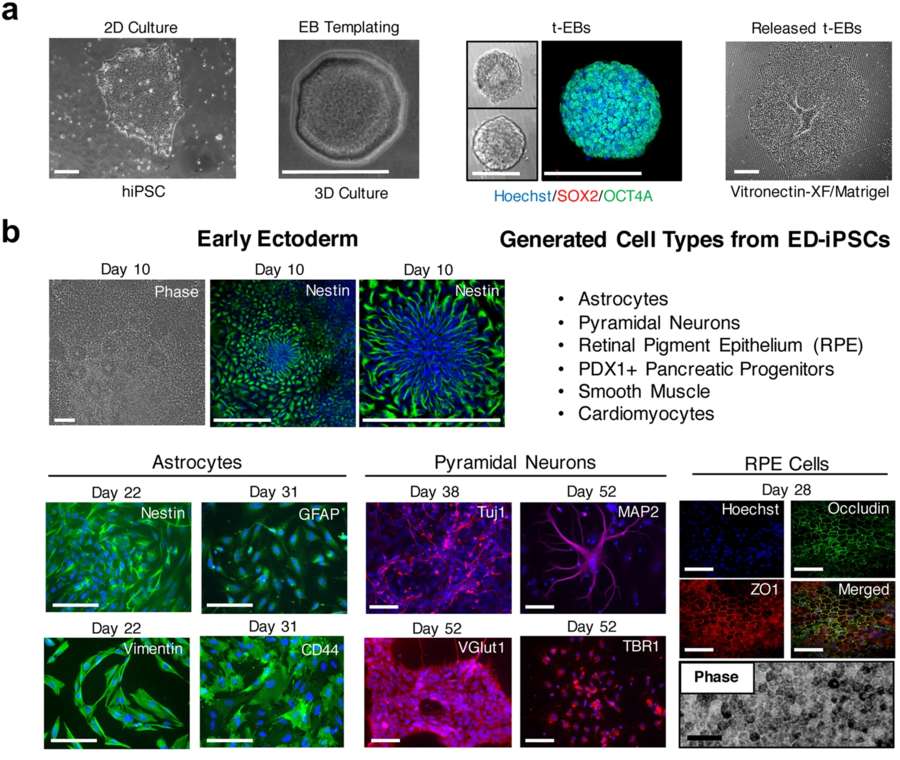 Fig. 1 Overview of differentiation strategies applied to ED-iPSC lines.1,2
Fig. 1 Overview of differentiation strategies applied to ED-iPSC lines.1,2
| Lineage Type | Derived Cell Types | Applications |
|---|---|---|
| Neuroectoderm | Neural progenitors, neurons, glia | Neuroscience, neurotoxicity, disease modeling |
| Surface ectoderm | Keratinocytes, lens cells | Dermatology, ocular biology, wound healing |
| Neural crest | Melanocytes, Schwann cells, craniofacial cells | Pigmentation studies, peripheral neuropathy, craniofacial research |
Efficient ectoderm differentiation depends on multiple factors including the quality of starting iPSCs, timing and duration of small molecule application, and downstream lineage-specific cues. Optimized protocols consistently yield over 80% purity of neuroectodermal populations.
The ability to generate ectodermal derivatives from iPSCs opens new frontiers in regenerative medicine, neuropharmacology, dermatological testing, and developmental toxicology. Our ectoderm differentiation protocols are finely tuned for reproducibility and scalability, ensuring robust yields and minimal batch variability. Our iPSC-to-ectoderm solutions can be tailored to your project's specific needs.
| Item | Specification |
|---|---|
| Cells and culture reagents |
|
| Differentiation reagents |
|
| Other supplies |
|
Seed iPSCs onto Matrigel-coated plates using mTeSR1. Change medium daily and expand to ~80% confluency before initiating differentiation. Use only healthy, undifferentiated colonies with defined borders.

Switch to ectoderm induction medium. Maintain induction for 5–7 days, changing media daily. Observe morphological changes: flattened cells with rosette-like clusters (neuroectoderm).

Based on the downstream lineage desired, introduce further patterning cues. For neuroectoderm: Maintain in dual SMAD inhibition medium. For neural crest cells: Add CHIR99021 to induce Wnt activation, supplement with FGF2 for neural crest survival. For surface ectoderm: Withdraw LDN and use BMP4 with low FGF signaling to promote non-neural ectoderm.
Successful differentiation of human iPSCs into ectodermal lineages is characterized by a series of predictable morphological, molecular, and functional outcomes. The quality and identity of the ectodermal derivatives can be confirmed by a combination of phase-contrast microscopy, immunocytochemistry, flow cytometry, and gene expression profiling.
| Lineage | Positive Markers | Negative Controls |
|---|---|---|
| Neuroectoderm | SOX1, PAX6, NESTIN, ZIC1 | OCT4, Brachyury, SOX17 |
| Neural crest | SOX10, p75 (NGFR), HNK-1, FOXD3 | PAX6, TP63 |
| Surface ectoderm | TP63, KRT18, KRT8 | SOX1, NESTIN, SOX17 |
Immunostaining reveals >80% positive staining for lineage-specific markers under optimal conditions. RT-qPCR demonstrates significant upregulation (≥10-fold) of lineage genes compared to undifferentiated iPSCs. Flow cytometry yields high-purity populations when patterning steps are correctly implemented.
| Problem | Possible Cause | Solution |
|---|---|---|
| Low differentiation efficiency |
|
|
| Cell detachment or death during induction |
|
|
| Heterogeneous populations |
|
|
| Spontaneous differentiation before induction |
|
|
| Poor neural rosette formation |
|
|
Creative Biolabs understands the complexity of iPSC differentiation workflows. Our team offers tailored optimization services including:
Creative Biolabs offers an end-to-end portfolio of iPSC-related solutions tailored to support research, screening, and preclinical modeling across multiple disciplines.
We offer high-efficiency reprogramming services using non-integrating methods to generate patient-specific or healthy donor-derived iPSCs.
Our gene editing platform supports targeted modifications using CRISPR/Cas9 technologies with high on-target precision.
We provide robust and lineage-specific differentiation protocols for generating a wide range of functional cell types from iPSCs.
Integrate iPSC-derived ectodermal cells into advanced 3D models or disease-relevant co-culture systems.
Whether you're exploring early development pathways, modeling neurological disorders, or seeking robust in vitro systems for screening, our iPSC-derived ectodermal solutions are built to accelerate your innovation.
Contact our iPSC specialists today to discuss your specific needs or request a custom quote.
References
Created July 2025
For Research Use Only. Not For Clinical Use.