Karyotype analysis is a fundamental cytogenetic tool to evaluate chromosomal stability and structural integrity in stem cells. Creative Biolabs specializes in high-quality stem cell characterization services, including comprehensive karyotyping assays, designed to help researchers validate the safety and consistency of their stem cell lines.
In this protocol, we detail the step-by-step workflow for karyotype analysis of stem cells, including the rationale, required materials, procedures, and troubleshooting.
Karyotyping is a classic cytogenetic technique that enables researchers to visualize the full set of chromosomes within a cell. In stem cell research, this method is indispensable because chromosomal integrity directly reflects the genomic stability and safety profile of the cell population. Even subtle chromosomal aberrations may alter differentiation potential, bias experimental outcomes, or pose risks in translational applications.
Karyotyping is based on visualizing metaphase chromosomes under a microscope after cells have been arrested in mitosis. Chromosomes are stained to produce banding patterns that enable identification of structural and numerical abnormalities.
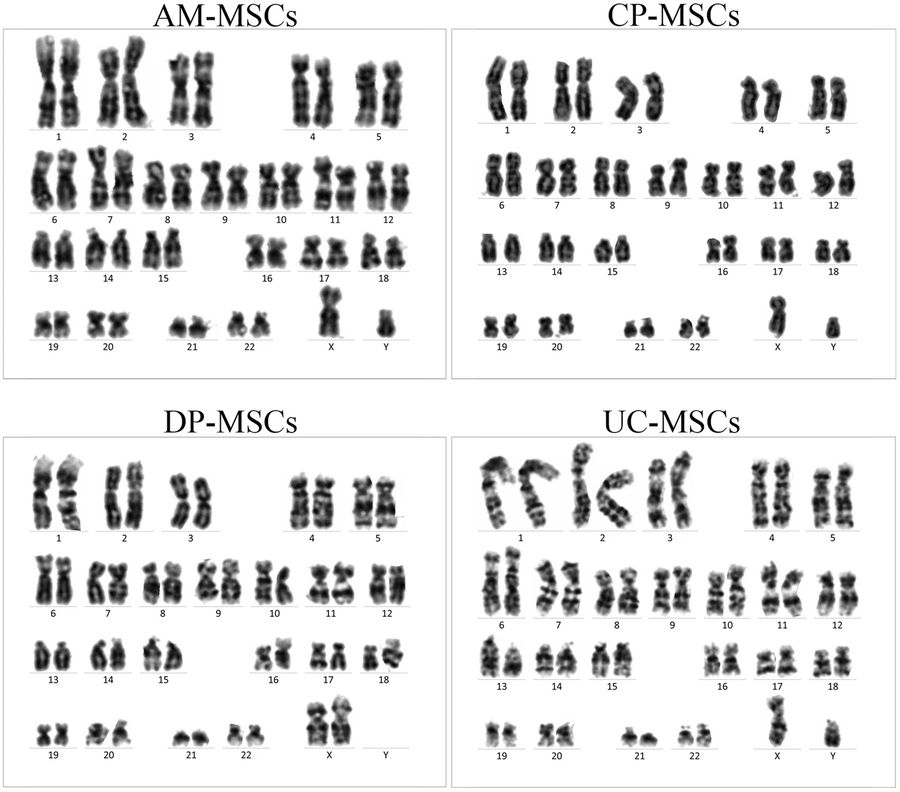 Fig.1 Karyotype analysis of different MSCs.1,2
Fig.1 Karyotype analysis of different MSCs.1,2
This approach allows detection of aneuploidies, translocations, inversions, duplications, and deletions, providing critical information about stem cell genomic integrity.
At Creative Biolabs, we have optimized each of these steps using advanced cytogenetic platforms and digital imaging systems. This ensures not only accurate detection of chromosomal anomalies but also reproducible, publication-quality results for your stem cell projects.
| Category | Item |
|---|---|
| Stem Cell Cultures | ESCs, iPSCs, MSCs |
| Mitotic Arrest Reagents |
Colcemid (demecolcine) Alternatives: Nocodazole, Colchicine |
| Hypotonic Solution |
0.075 M KCl Alternatives: Sodium citrate, diluted KCl |
| Fixatives | Methanol: Acetic Acid (3:1) |
| Staining Reagents |
Giemsa stain Alternative banding: GTG, Q-banding, C-banding |
| Cell Handling Reagents |
Trypsin-EDTA solution PBS Complete culture medium |
Culture stem cells under optimal conditions until they reach ~70–80% confluence. Ensure cells are actively dividing, as non-dividing cells will not contribute to metaphase spreads. Avoid excessive passaging prior to analysis, as this may alter karyotypic profiles.

Add colcemid to the culture medium. Incubate cells for 1–2 hour, depending on cell type. Monitor under the microscope for increased mitotic figures.

Detach cells using trypsin-EDTA solution. Neutralize with culture medium containing serum. Centrifuge and discard supernatant and gently resuspend the cell pellet in PBS.

Add pre-warmed 0.075 M KCl hypotonic solution to the pellet. Incubate at 37 °C for 20 minutes. This step swells cells, spreading chromosomes apart for clear visualization.

Carefully add ice-cold fixative (methanol: acetic acid, 3:1). Mix gently and centrifuge. Repeat fixation 2–3 times to ensure clean chromosomal spreads.

Drop cell suspension onto chilled microscope slides from a height of 30–50 cm to encourage chromosome spreading. Allow slides to air-dry at room temperature.

Stain slides with Giemsa for 5–10 minutes. Rinse briefly with distilled water. Air-dry and mount with coverslips using DPX mounting medium.

Examine under a brightfield microscope. Capture digital images of well-spread metaphase cells. Prepare karyograms by arranging homologous chromosomes into pairs. Analyze for structural and numerical abnormalities.
Below is a comprehensive troubleshooting guide to help you achieve reliable and high-quality results.
| Problem | Possible Cause | Solution |
|---|---|---|
| Low mitotic index (few metaphases visible) |
|
|
| Poor metaphase spreads (chromosomes clumped together) |
|
|
| Chromosome loss or fragmented chromosomes |
|
|
| Weak or inconsistent banding patterns |
|
|
| Background debris obscuring chromosomes |
|
|
| Overlapping chromosomes in most spreads |
|
|
| Recurrent chromosomal abnormalities across passages |
|
|
To provide our clients with comprehensive genomic and functional insights, we offer an integrated portfolio of supporting services tailored to stem cell research and translational applications.
Flow cytometry, qPCR, and immunostaining assays to validate stemness markers (OCT4, SOX2, NANOG) and confirm differentiation capacity.
Generation of high-quality human iPSCs from somatic cells using non-integrative methods.
Development of lineage-specific differentiation protocols for disease modeling, drug testing, or cell therapy research.
Gene knock-out, knock-in, or correction services for iPSC lines using precise strategies with clone validation.
A: Best practice is to perform karyotyping at early passages, before major manipulations such as gene editing, and at routine intervals during long-term culture (e.g., every 10 passages). This strategy helps detect genomic instability early and prevents investing resources in compromised cell populations.
A: No. Karyotyping is excellent for detecting large chromosomal changes such as aneuploidy, translocations, or deletions above ~5–10 Mb. Smaller genetic alterations, including point mutations or microdeletions, require complementary methods such as SNP arrays, FISH, or next-generation sequencing for comprehensive genomic assessment.
A: Recurrent abnormalities include trisomy 12, trisomy 8, and isochromosome 17q in human pluripotent stem cells. These changes can influence proliferation and differentiation. Detecting such anomalies early allows researchers to discontinue unstable lines and maintain only high-quality, genetically intact cells for downstream use.
A: Giemsa staining (G-banding) remains the gold standard due to its reproducibility and clarity. However, advanced banding techniques such as Q-banding or C-banding can be employed for specific applications. At Creative Biolabs, we tailor staining strategies to the unique needs of your stem cell line and research goals.
References
Created August 2025
For Research Use Only. Not For Clinical Use.