Creative Biolabs understands that accuracy in HLA typing translates to reliability in stem cell applications. Our HLA typing analysis protocol offers a robust and reproducible workflow, balancing molecular precision with practical feasibility. This protocol is designed for researchers aiming to perform reliable HLA typing on stem cell populations and highlights common challenges, optimization strategies, and service-related solutions.
The principle of HLA typing lies in the identification of polymorphic sequences within the Major Histocompatibility Complex (MHC) on chromosome 6. These polymorphisms define an individual's HLA alleles, which in turn govern immune recognition.
HLA genes encode glycoproteins that present antigenic peptides to T cells. The extreme polymorphism of these genes, especially within the HLA class I (HLA-A, -B, -C) and class II (HLA-DR, -DQ, -DP) loci, forms the immunological identity of every human.
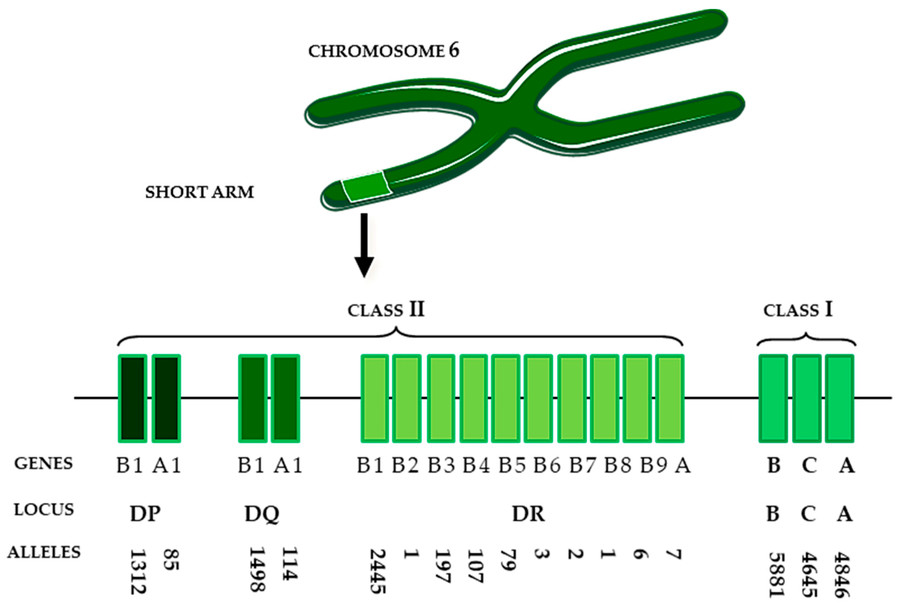 Fig.1 HLA region on chromosome 6.1,2
Fig.1 HLA region on chromosome 6.1,2
In stem cell research, mismatched HLA alleles can provoke graft-versus-host disease (GvHD) or graft rejection. Therefore, precise HLA typing safeguards therapeutic safety and efficacy.
| Category | Item |
|---|---|
| Stem Cell Source |
Hematopoietic stem cells (HSCs): From bone marrow, cord blood, or peripheral mobilized blood. Mesenchymal stem cells (MSCs): Derived from bone marrow, adipose tissue, or iPSCs. Induced pluripotent stem cells (iPSCs): Derived from somatic cell reprogramming. |
| DNA Extraction Reagents |
Cell lysis buffer Proteinase K RNase A Ethanol (molecular grade) Spin columns or magnetic bead-based DNA purification kits |
| PCR Components |
Taq DNA polymerase (high fidelity) dNTP mix PCR buffer with MgCl₂ Sequence-specific primers (for PCR-SSP) Sequence-specific oligonucleotide probes (for PCR-SSO) |
| Sequencing Reagents |
Sanger sequencing kits Capillary electrophoresis consumables NGS library preparation kits Barcoded adapters and primers for multiplexing |
Harvest stem cells under sterile conditions. Isolate genomic DNA using spin-column or magnetic bead-based purification. Quantify DNA.

Amplify exons 2 and 3 (for class I genes) and exon 2 (for class II genes). Validate amplification via agarose gel electrophoresis.

PCR-SSP: Quick, cost-effective, low resolution. PCR-SSO: High-throughput, medium resolution. SBT: High resolution, labor-intensive. NGS: Comprehensive, scalable, and most accurate.

For Sanger sequencing, perform cycle sequencing and capillary electrophoresis. For NGS, prepare indexed libraries, sequence, and perform bioinformatics alignment.

Compare sequences against the IMGT/HLA database. Assign alleles at 2-digit (low), 4-digit (intermediate), or 6-digit (high) resolution.
Below, we provide a guide to common issues, their underlying causes, and practical optimization strategies. These tips are derived from Creative Biolabs' extensive hands-on experience in stem cell characterization projects.
| Problem | Possible Cause | Solution |
|---|---|---|
| Low DNA yield or poor DNA quality |
|
|
| PCR amplification failure |
|
|
| Non-specific amplification or background bands |
|
|
| Sequencing artifacts |
|
|
| Ambiguous allele calls |
|
|
To support researchers and biotech innovators, we provide a comprehensive ecosystem of services that align seamlessly with HLA typing analysis. Our integrated portfolio ensures your project advances with confidence.
A: NGS is currently the gold standard, delivering allele-level resolution across multiple loci. Compared with PCR-based techniques, NGS provides comprehensive coverage, reduces allele ambiguity, and scales easily for large cohorts. It is ideal for stem cell banks, clinical-grade stem cell therapies, and translational immunogenetics research.
A: Common issues include low DNA yield, ambiguous allele assignments, and sequencing artifacts from heterozygous loci. These challenges can be overcome by optimizing DNA extraction, using high-fidelity polymerases, and employing NGS platforms. At Creative Biolabs, our troubleshooting protocols guarantee clean, reproducible datasets.
A: Yes. With NGS platforms, we routinely identify rare or novel alleles absent in conventional typing panels. These findings can be submitted to international databases for validation. Our team ensures such cases are confirmed with repeat sequencing, providing clients with accurate, publication-ready data.
A: Yes. Genomic DNA from iPSCs is highly suitable for HLA typing. Typing iPSCs ensures compatibility for downstream differentiation, transplantation, and universal donor cell line development. For global stem cell banks, this step guarantees that each iPSC line has a standardized, traceable immunogenetic identity.
References
Created August 2025
For Research Use Only. Not For Clinical Use.