The ability to reprogram somatic cells into induced pluripotent stem cells (iPSCs) has transformed regenerative medicine and stem cell research. Among the different strategies, RNA-based reprogramming has emerged as a powerful, non-integrating, and highly efficient method. By avoiding permanent genomic integration, RNA-driven approaches ensure safer and more controllable generation of iPSCs for disease modeling, drug screening, and cell-based therapy development.
At Creative Biolabs, we have dedicated years of expertise to refining RNA-based reprogramming protocols, enabling our clients to accelerate research projects with precision and reproducibility. Below, we outline a comprehensive, step-by-step protocol enriched with practical insights from our laboratory practice.
RNA-based reprogramming rests on several scientific and technical advantages that make it particularly attractive compared to DNA-based or viral-vector methods. Unlike retroviral or lentiviral methods, which integrate transcription factors into the host genome, RNA-based systems deliver mRNAs or self-replicating RNAs that remain episomal. This transient expression avoids permanent genomic modifications and significantly lowers the risk of insertional mutagenesis or oncogenic transformation.
Synthetic modified mRNAs can achieve robust protein expression within hours, bypassing transcriptional regulation steps. This direct cytoplasmic translation enables stronger and faster induction of pluripotency genes compared to DNA vectors. In addition, daily delivery maintains steady protein levels, producing high reprogramming efficiency across different somatic cell types.
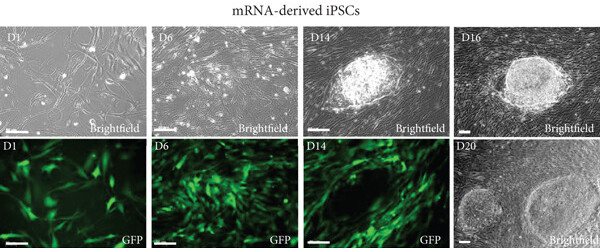 Fig.1 mRNA-based iPSC reprogramming of fibroblasts.1,2
Fig.1 mRNA-based iPSC reprogramming of fibroblasts.1,2
RNA-based reprogramming is not restricted to fibroblasts. It has been successfully applied to keratinocytes, blood cells, and even urine-derived epithelial cells. This flexibility broadens the accessibility of iPSC generation, making it suitable for diverse research applications ranging from rare genetic diseases to large-scale drug screening.
Because RNA reprogramming is feeder-free and xeno-free compatible, it lends itself to standardized culture conditions. Synthetic mRNAs can be manufactured under GMP guidelines, and the workflow can be automated with robotic liquid handlers. These features allow for scalable, reproducible iPSC production suitable for CRO environments and industrial pipelines.
| Category | Reagents |
|---|---|
| Somatic cells | Dermal fibroblasts (primary or immortalized) |
| Modified mRNAs | Synthetic, pseudouridine- and 5-methylcytidine-modified transcripts encoding OCT4, SOX2, KLF4, c-MYC, LIN28, and NANOG |
| RNA delivery reagent | Lipid-based transfection reagent optimized for mRNA |
| Cell culture media |
DMEM with high glucose (for fibroblast expansion) Reprogramming medium (mTeSR1 or equivalent feeder-free medium) |
| Supplementary factors |
B18R recombinant protein (type I interferon inhibitor) Small molecules (e.g., valproic acid, CHIR99021, SB431542) |
| Matrix coating | Matrigel or vitronectin for feeder-free iPSC culture |
Plate fibroblasts onto Matrigel-coated 6-well plates. Grow in fibroblast medium until 80% confluence. Confirm absence of mycoplasma contamination. Ensure viability >90% by trypan blue exclusion. Replace fibroblast medium with reprogramming medium supplemented with B18R protein 24 hours before RNA transfection.

Combine equal molar amounts of OCT4, SOX2, KLF4, c-MYC, LIN28, and NANOG mRNAs. Adjust concentration according to transfection reagent specifications. Dilute mRNA mix in Opti-MEM (serum-free medium). Mix with lipid-based transfection reagent and incubate at room temperature. Add the transfection complex dropwise to cells. Replace media with fresh reprogramming medium containing B18R after 4–6 hours. Repeat transfection daily for 10–12 days.

Replace medium every 24 hours with fresh reprogramming medium plus small molecules. Day 3–5: Cells become more compact with high nuclear-to-cytoplasmic ratio. Day 7–10: Emergence of epithelial-like colonies with clear borders. Add small molecules to enhance reprogramming efficiency. Avoid over-confluence by splitting if necessary. Watch for cell death—optimize RNA dosage if survival is <50%.

Around day 12–18, colonies with defined borders and prominent nucleoli appear. Use live staining (e.g., TRA-1-60 or SSEA-4) for confirmation. Manually pick individual colonies under a stereomicroscope. Transfer colonies to fresh Matrigel-coated wells with mTeSR1 medium. Passage colonies every 5–6 days. Maintain feeder-free conditions with daily medium changes.
Below we provide an expanded troubleshooting and optimization guide to help researchers maximize efficiency and reproducibility.
| Problem | Possible Cause | Solution |
|---|---|---|
| Low transfection efficiency |
|
|
| High cell death after transfection |
|
|
| Few or no iPSC colonies form |
|
|
| Colonies appear but differentiate rapidly |
|
|
| Genetic instability in iPSCs |
|
|
Our RNA-based stem cell reprogramming protocol is part of a much broader portfolio designed to accelerate your stem cell projects. Each service is carefully optimized to complement reprogramming workflows, providing you with end-to-end support from the earliest stages of somatic cell preparation to advanced functional assays.
Our team specializes in generating high-quality iPSCs using multiple approaches, including RNA-based, episomal vector, and Sendai virus methods. By offering a spectrum of technologies, we can tailor reprogramming to your research requirements, ensuring safety, reproducibility, and efficiency.
We provide a full suite of QC assays, including immunofluorescence for pluripotency markers, gene expression profiling, epigenetic state analysis, karyotyping, and embryoid body formation tests. This ensures that your iPSCs meet the highest standards of genetic stability and differentiation potential.
We offer customized differentiation protocols to generate lineage-specific cell types, such as cardiomyocytes, hepatocytes, neural progenitors, and pancreatic β-like cells.
For researchers interested in gene-function studies or disease modeling, our genome editing platform integrates seamlessly with iPSC technology.
With over 20 years of experience as a trusted CRO partner, Creative Biolabs is committed to advancing stem cell science and providing reliable solutions tailored to your research goals.
A: Fibroblasts remain the most common donor cell type, but RNA-based reprogramming is also effective with keratinocytes, blood cells, and even urine-derived epithelial cells. At Creative Biolabs, we adapt our RNA cocktails and delivery systems to match the biology of your chosen donor cell, ensuring that your project begins with the highest possible efficiency.
A: Most projects require 12–18 days from the initial RNA transfection to the appearance of well-defined iPSC colonies. Early morphological changes can be seen as soon as Day 3–5, with epithelial-like colonies forming by Day 7–10. Creative Biolabs provides detailed monitoring timelines and real-time QC checks to ensure your colonies are picked and expanded at the optimal stage.
A: Yes. Depending on the research objective, we can customize RNA cocktails by adding or removing factors such as LIN28 or NANOG, or by incorporating lineage-specific transcriptional regulators. We also adjust small-molecule supplementation and culture conditions to enhance efficiency for challenging donor cell types.
A: Beyond reprogramming, we offer directed differentiation into multiple lineages, genome editing, and advanced 3D organoid development. By partnering with us, you gain access to an end-to-end platform that transforms somatic cells into iPSCs and then into functional models, with robust QC and regulatory guidance at each stage.
References
Created September 2025
For Research Use Only. Not For Clinical Use.