Fluorophore Modification Service
Introduction
Fluorophores are luminescent molecules that can absorb and re-emit light at specific wavelengths and change after binding with oligonucleotides. They can track in real time, conduct quantitative analysis, and perform high-resolution imaging of nucleic acids, which is crucial for FISH, live cell research, diagnostic analysis, and validation of target participation in treatment.
At Creative Biolabs, we offer end-to-end fluorophore-conjugated oligonucleotide solutions: customizing the attachment of various fluorophores (FAM, Cy3, Cy5, etc.) through proprietary chemical methods; High-purity synthesis by HPLC/MS, and probes tailored for imaging, diagnosis, or treatment monitoring. With over 20 years of professional expertise, our tools combine precision and reliability to accelerate your research.
[Contact Our Experts for Tailored Solutions]
Fluorophores
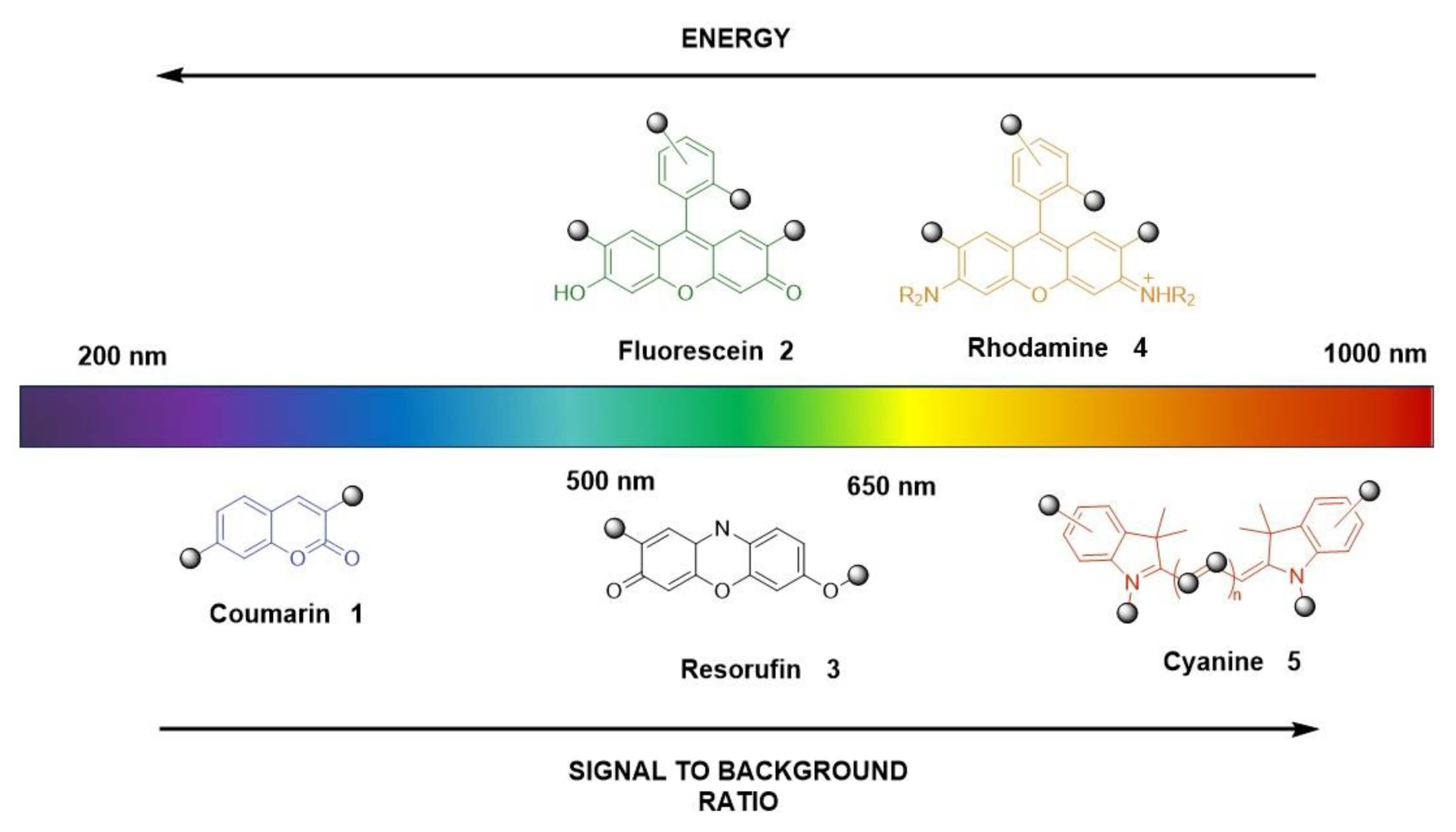 Fig.1 The most common fluorophores summarized by structure and emission color.1
Fig.1 The most common fluorophores summarized by structure and emission color.1
Basic Properties
Fluorophores are light-emitting molecules covalently conjugated to oligonucleotides at specific positions, 5', 3', or internal bases, enabling real-time visualization of cellular uptake, trafficking, and target interactions. Common options include Cyanine dyes (Cy3, Cy5) and fluorescein derivatives, optimized to minimize interference with oligonucleotide function.
Key Advantages
Their key advantages include enhanced detection sensitivity (supporting sub-nanomolar tracking in live-cell imaging or in vivo biodistribution), dual functionality (combining therapeutic activity like gene silencing with diagnostic capabilities such as tumor targeting validation), and reduced off-target effects (site-specific labeling preserves base-pairing specificity and limits steric hindrance).
Design Considerations
For effective design, focus on labeling position (terminal modifications at 3'/5' optimize signal without disrupting hybridization), quenching mitigation (avoid placing fluorophores near G-quadruplex regions or nucleases to prevent signal loss), and linker chemistry (use non-polar, flexible linkers like C6 or PEG spacers to reduce conformational strain).
Workflow
-
Sequence Design & Fluorophore Selection
Share your target sequence and preferred fluorophore (such as Cy5 for in vivo imaging needs), and our team of experts will step in to optimize labeling sites. This careful adjustment ensures a balance between the oligonucleotide's stability and the strength of the fluorescent signal, laying a solid foundation for subsequent experiments.
-
Synthesis & Conjugation
We employ automated phosphoramidite chemistry to achieve precise incorporation of fluorophores during solid-phase synthesis, ensuring each modification is accurately positioned. After synthesis, post-synthesis HPLC purification is performed to thoroughly remove any unbound fluorophores, guaranteeing the purity of the conjugated oligonucleotides.
-
Quality Control
Our rigorous quality control process covers two key aspects: purity verification, through IP-HPLC and MALDI-TOF analysis; and functionality validation, which includes testing fluorescence intensity and confirming the target binding affinity, ensuring the modified oligonucleotides perform as expected.
-
Delivery
You will receive lyophilized oligonucleotides along with comprehensive reports, including HPLC traces and mass spectrometry data, within 4-8 weeks. This timely delivery, paired with detailed documentation, supports a smooth transition to your experimental workflow.
Request a Customized Project Timeline
What We Can Offer?
Tailored Fluorophore Integration
Our proprietary algorithms identify optimal labeling sites by analyzing sequence structure and application needs, maximizing signal-to-noise ratios while avoiding interference with oligonucleotide function, ideal for sensitive assays.
Ultra-High Purity
HPLC and PAGE purification eliminate impurities like diastereomers and free fluorophores, ensuring purity for consistent results in quantitative tests.
Scalability
We support 10 nmol to 10 mmol synthesis under uniform quality standards, enabling smooth transitions from early R&D to large-scale production.
Multi-Modality Support
Seamlessly combine fluorophores with phosphorothioate or 2'-O-methyl modifications to boost stability and bioavailability, perfect for long-term in vivo tracking or theragnostic.
Rapid Turnaround
Expedited service delivers high-quality modified oligonucleotides in 3-5 weeks, with dedicated support to meet tight project deadlines.
[Legare Our Cutting-Edge Oligonucleotide Platform – Schedule a Consultation]
Customer Reviews

FAQs
Q: How does the choice of linker (e.g., C6 vs. PEG) impact fluorophore performance in oligonucleotides?
A: Linker type affects flexibility and solubility. C6 linkers enhance stability in organic solvents (ideal for in vitro assays), while PEG linkers improve aqueous solubility, critical for in vivo applications, reducing non-specific tissue binding by 30-40% in our validation data.
Q: What is the maximum number of fluorophores that can be conjugated to a single oligonucleotide without causing aggregation?
A: Typically 2-3 fluorophores for 20-30 nt oligonucleotides. Higher density increases hydrophobicity, risking aggregation. Our algorithms predict optimal stoichiometry based on sequence length and fluorophore size.
Q: How do you mitigate fluorescence quenching caused by G-rich sequences in oligonucleotides?
A: We insert a basic spacer between fluorophores and G-quadruplex-forming regions, reducing quenching by 50-70%. Post-synthesis fluorescence spectroscopy confirms signal retention.
Q: For long-term in vivo tracking, do fluorophores affect oligonucleotide clearance rates?
A: Minimal impact when using hydrophilic fluorophores. Our data shows clearance half-lives for modified oligonucleotides differ by <10% from unmodified versions, with no accumulation in major organs.
[Contact Our Team for More Information and to Discuss Your Project]
Reference
- Gil-Rivas, Alba, et al. "New advances in the exploration of esterases with PET and fluorescent probes." Molecules 28.17 (2023): 6265. DOI: 10.3390/molecules28176265. Distributed under Open Access license CC BY 4.0, without modification.
