Identity Service of Nucleic Acid
Nucleic acids are intricate macromolecules composed of nucleotide monomers, forming elongated chains. Therapeutics and vaccines based on nucleic acids encompass a variety of forms including mRNA, siRNA, and antisense oligonucleotides (ASOs). These modalities exhibit numerous exceptional therapeutic attributes such as remarkable specificity, functional versatility, and straightforward adaptability. Characterization and analysis of nucleic acids are pivotal in drug development, regulatory approval, and quality assurance, involving the identification of nucleic acid type, sequence, and any modifications or variations present. Creative Biolabs employs diverse approaches, including molecular weight determination and sequencing, to ensure the accuracy of oligonucleotide identity. Molecular weight and sequencing serve as essential verification methods.
Nucleic Acid Introduction
Nucleic acids, including deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), are the core information molecules of all known life forms. They are biopolymers, essential for all life forms, and their primary function is to store and express genetic information. Structurally, nucleic acids are long chains composed of monomeric units called nucleotides. Each nucleotide consists of three parts: a nitrogenous base, a pentose sugar (deoxyribose in DNA and ribose in RNA), and a phosphate group. The sugar-phosphate backbone forms the structural backbone of the molecule, while the sequence of nitrogenous bases (adenine (A), guanine (G), cytosine (C), and thymine (T) in DNA, and uracil (U) in RNA) encodes genetic instructions.
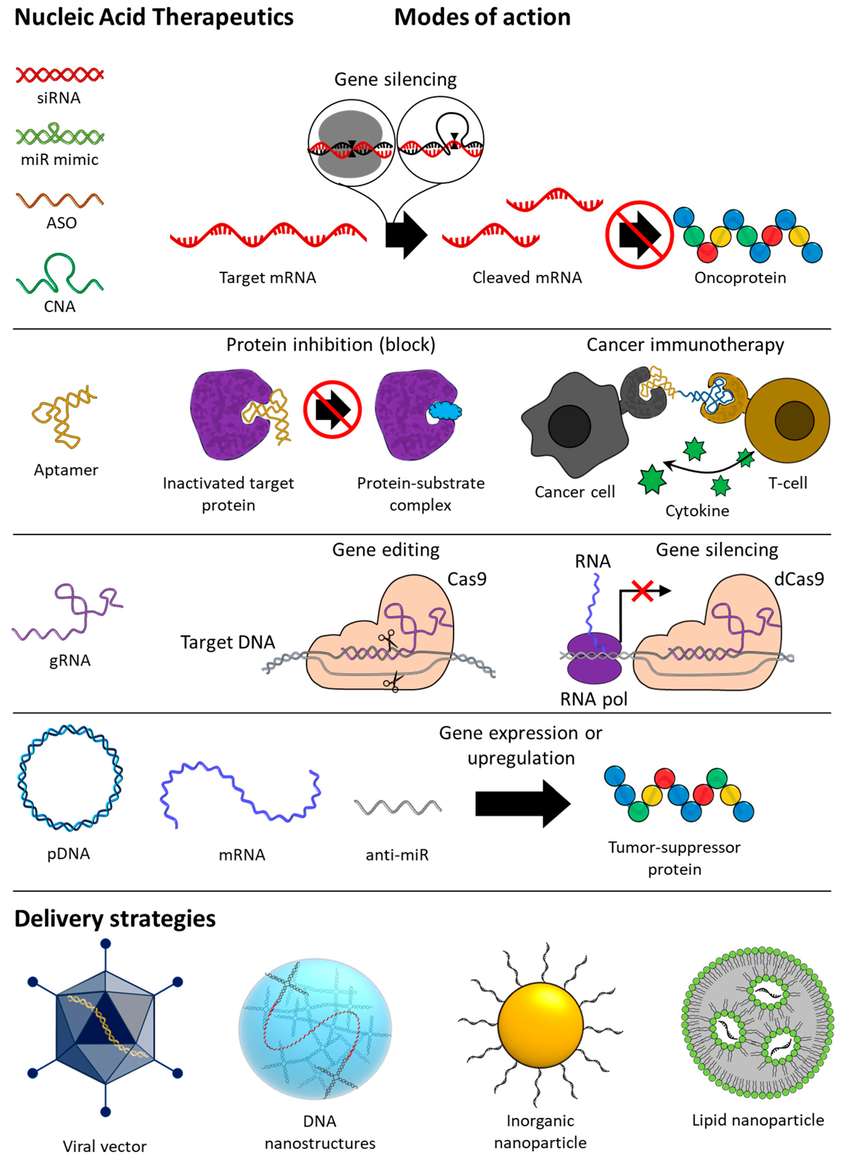 Figure 1. Overview of types, modes of action, and delivery strategies of nucleic acid-based therapeutics for anti-cancer therapy. siRNA, small interfering RNA; miR, microRNA; ASO, antisense oligonucleotide; CNA, catalytic nucleic acid; gRNA, guide RNA; pDNA, plasmid DNA; mRNA, messenger RNA.1
Figure 1. Overview of types, modes of action, and delivery strategies of nucleic acid-based therapeutics for anti-cancer therapy. siRNA, small interfering RNA; miR, microRNA; ASO, antisense oligonucleotide; CNA, catalytic nucleic acid; gRNA, guide RNA; pDNA, plasmid DNA; mRNA, messenger RNA.1
Identification of Nucleic Acid
Nucleic acid identification is a complex process that cannot be accomplished with simple testing. It involves qualitative and quantitative assessments to determine the presence, purity, concentration, integrity, and specific sequences of DNA or RNA molecules in a biological sample. The complexity of this task stems from several factors: the target substance is often present in extremely low concentrations; the need to distinguish highly similar sequences (e.g., differentiating between somatic mutations and germline variations); and the presence of nucleic acids in a wide variety of biological matrices (blood, tissue, cell culture supernatant).
Technology of Identification Service of Nucleic Acid
The technological system for nucleic acid identification has undergone profound changes, shifting from gel-based separation methods to high-throughput, massively parallel sequencing platforms.
Table.1 Several approaches for identifying nucleic acids.
| Test Methods | Description | Application |
|---|---|---|
| Mass Spectrometry (MS) | MS involves ionizing compounds to create charged molecules or fragments, which are then separated by mass-to-charge ratio (m/z) and detected. | Qualitative analysis of oligo samples to determine the absence of target sequence. |
| Liquid Chromatography with tandem mass spectrometry (LC-MS-MS) |
MS sequencing ensures the accuracy of oligo sequence arrangement and modification by matching production information with theoretical sequence product ions. This powerful analytical approach integrates the separation abilities of liquid chromatography with the outstanding sensitivity and selectivity of triple quadrupole mass spectrometry. It remains invaluable for validating sequences and detecting modifications. |
Validate the sequence of nucleic acids. By separating and ionizing nucleic acid samples, and then analyzing the resulting ion mass-to-charge ratio (m/z) with a mass spectrometer, the base sequence of the nucleic acid can be determined. Detect and identify modification groups in nucleic acids, such as methylation, phosphorylation, etc. This is crucial for studying the function and metabolism of nucleic acids. Detect and identify hybridizations between nucleic acids, such as in RNAi (RNA interference) studies, to determine the hybridization of siRNA to mRNA. |
| Sanger Sequencing |
It is particularly valuable for detecting and determining the nucleotide sequences of both mRNA and plasmid DNA. Renowned for its high accuracy and reliability, it is ideal for sequencing single genes or small DNA segments. |
Identify the precise sequence of nucleotides in mRNA molecules. Confirm the integrity of plasmid constructs, verify the presence of desired DNA inserts, and ensure accurate genetic engineering. |
| Next-Generation Sequencing (NGS) | It is a high-throughput technology used for nucleic acid sequencing. It rapidly and accurately determines the sequences of DNA or RNA, generating large amounts of data at a relatively low cost. | Detect both single-stranded and double-stranded DNA as well as RNA sequences. |
| Third generation sequencing | Characterized by their capability to directly sequence single DNA molecules in real-time, without relying on PCR amplification or DNA fragment cloning, technologies such as Single-Molecule Real-Time (SMRT) and Oxford Nanopore sequencing are prominent examples. |
Detect both single-stranded and double-stranded DNA, RNA sequences, or sequence libraries within target samples. It is particularly effective for long sequences or those containing repetitive elements. Additionally, it enables the sequencing of long single-stranded DNA molecules. |
Applications of Nucleic Acid
01 Target Identification and Validation
Identifying differentially expressed nucleic acid sequences (e.g., mRNA transcripts or non-coding RNA) in disease and healthy states is the first step in discovering new drug targets.
02 Biomarker Discovery
Specific DNA mutations, gene copy number variations, or altered RNA expression profiles can serve as molecular biomarkers for early disease diagnosis, prognosis, and predictive treatment response.
03 Epigenetics
Identification services cover chemically modified nucleic acids, such as DNA methylation profiling using techniques like whole-genome bisulfite sequencing (WGBS) or reduced representative bisulfite sequencing (RRBS).
04 Pathogen Identification
Rapid and accurate identification of microbial DNA or RNA is crucial for controlling infectious disease outbreaks and guiding antibiotic/antiviral therapy.
Identify the Nucleotides Which Contain Purines in a Nucleic Acid Strand
The specific identification of purine nucleotide bases in nucleic acid chains is a unique analytical challenge of significant biological importance. Purine bases (adenine and guanine) and their modified derivatives play crucial roles in the structure, function, and recognition of nucleic acids. Their unique chemical properties enable specific detection and characterization using a variety of analytical methods.
Chemical methods for purine identification utilize the difference in the sensitivity of glycosidic bonds to acid hydrolysis. Under controlled acidic conditions, the glycosidic bonds of purine nucleotides are more readily hydrolyzed than those of pyrimidine nucleotides, resulting in selective depurination. The released purine bases can then be separated and quantified using chromatographic techniques, providing information on purine composition and relative abundance.
Our Services
Our organization provides comprehensive nucleic acid identification services leveraging state-of-the-art technological platforms and extensive analytical expertise. We offer customized solutions addressing diverse research needs across academic, pharmaceutical, biotechnology, and clinical sectors. Our service portfolio encompasses multiple levels of nucleic acid analysis, from basic characterization to advanced functional assessment.
Why Choose Our Services?
Technological Excellence
Our operations utilize state-of-the-art instruments and are maintained in strict accordance with quality control procedures. We continuously evaluate and apply the latest methodologies to ensure our services remain at the forefront of analytical capabilities.
Quality Assurance
We implement a comprehensive quality management system across all analytical processes. Our standardized operating procedures, regular calibrations, use of reference materials, and the implementation of proficiency testing programs ensure consistent performance and reliable results.
Customized Solutions
We understand that every research project has unique needs and challenges. Our service approach emphasizes collaborative consultation to develop optimized analytical strategies tailored to specific research objectives.
Result Delivery
- Raw Data Files: For sequencing projects, this includes FASTQ files containing raw sequence reads and quality scores.
- Statistical Analysis Report: A formal document detailing the methods used for statistical analysis, including normalization, filtering, and the final list of salient elements.
- Graphical Summary: High-resolution graphs, including volcano plots, heatmaps, principal component analysis (PCA) plots, and pathway enrichment plots, for immediate interpretation and use in presentations or publications.
- Quality Control Report: Detailed reports on nucleic acid isolation (concentration, purity, RIN/DIN values) and sequencing performance (read depth, alignment rate, quality score metrics).
Frequently Asked Questions
Q: How does Creative Biolabs ensure data quality and reproducibility of results?
A: We ensure data quality through three main mechanisms: 1) pre-sequencing quality control (purity/integrity checks), 2) process quality control (assessing library size distribution and molar concentration), and 3) post-sequencing quality control (ensuring minimum sequencing depth, alignment rate, and Q-value thresholds).
Q: Can you assist us in submitting our data to public databases such as GEO?
A: Absolutely. Our bioinformatics team is highly experienced in preparing data according to GEO standards. We help clients build metadata, normalize and format data tables, and generate necessary submission documents, thus streamlining their processes and ensuring their data meets open publication requirements.
Q: How long does a standard RNA-Seq project typically take?
A: The standard cycle from receiving samples to final data delivery and initial report generation is typically 6-8 weeks, depending on the complexity of the project and the required sequencing depth. We offer expedited services for time-sensitive projects.
Conclusion
Creative Biolabs provides top-tier nucleic acid identity services worldwide. Our services encompass determining nucleic acid sequences, identifying mutations or modifications, analyzing subtypes or splice variants, and elucidating RNA or DNA structure and interactions. For further details, reach out to us for a quotation. We guarantee a response within 24 hours and will tailor an optimal method for your project.
Reference
- Lee M, Lee M, Song Y, et al. Recent advances and prospects of nucleic acid therapeutics for anti-cancer therapy. Molecules, 2024, 29(19): 4737. https://doi.org/10.3390/molecules29194737 (Distributed under Open Access license CC BY 4.0, without modification.)
