Pseudotyping Service of Lentiviral Vectors with Gammaretrovirus MLV
Lentiviral vectors pseudotyped with the envelope glycoproteins of Moloney murine leukemia virus (MLV) represent a complementary molecular chassis that fuses the broad-host tropism of retroviral entry machinery with the superior cargo capacity and SIN safety profile of HIV-1-derived backbones. MLV amphotropic (4070A), ecotropic, and 10A1 envelopes retain a comparatively rigid post-fusion conformation and pH-independent membrane fusion kinetics, thereby conferring rapid, high-efficiency transduction of quiescent hematopoietic stem cells, naïve B-lymphocytes, and terminally differentiated airway epithelia that remain poorly accessible to VSV-G or BaEV-R pseudotypes. Recent engineering of MLV surface unit (SU) hypervariable regions—through alanine scanning, glycan shielding, and charge-reversal mutagenesis—has expanded the entry receptor repertoire from the classic PiT-1/PiT-2 phosphate transporters to include CD46 and CD134, while simultaneously abrogating cross-species transmission signatures. When these remodeled envelopes are co-packaged with third-generation lentiviral genomes carrying codon-optimized gag-pol, cPPT (central polypurine tract)/ CTS (central termination sequence) central polypurine tracts, and microRNA detargeting cassettes, the resulting vectors exhibit high titers, minimal Replication-Competent Lentivirus (RCL) formation, and integration profiles devoid of transcriptional hotspots. Consequently, MLV-pseudotyped lentiviral particles have become the vector of choice for lineage-tracing studies in primary human hematopoiesis, chimeric antigen receptor knock-in experiments under EF1α or PGK promoters exceeding 8 kb, and CRISPR base-editing screens in patient-derived airway organoids where restricted tropism and sustained expression are simultaneously required.
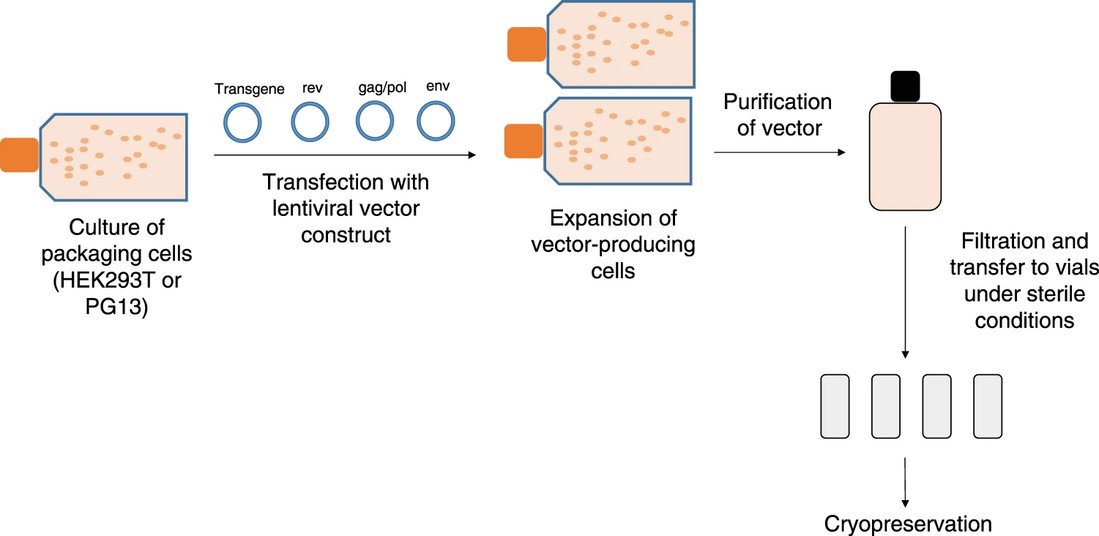 Fig. 1 Overview of large-scale vector manufacturing1,2
Fig. 1 Overview of large-scale vector manufacturing1,2
We understand that deploying lentiviral vectors into the most refractory cellular niches requires an envelope repertoire that transcends the conventional VSV-G paradigm. Creative Biolabs's Pseudotyping Service for MLV-Derived Envelopes unifies retroviral entry biology, structure-guided protein engineering, and high-resolution functional genomics into a turnkey platform, eliminating every discontinuity between your experimental question and a sequence-verified, MLV-pseudotyped lentiviral stock. Leveraging suspension-adapted HEK293T producer lines tuned for amphotropic, ecotropic, or 10A1 envelope variants, we deploy rapid mutagenesis libraries, glycan-shield mapping, and single-particle interferometry to engineer envelopes with defined receptor footprints and pH-independent fusion kinetics. The resulting particles are delivered as endotoxin-cleared, endothelial-free reagents ready for immediate transduction of different cells, polarized airway epithelia, or patient-derived organoids, propelling your mechanistic studies from conceptual design to publication-grade datasets without procedural latency.
Our Fast-Track Workflow
Creative Biolabs's MLV-pseudotyping suite fuses third-generation, suspension-adapted producer lines with envelope-specific codon harmonization and dual-mode membrane-affinity chromatography to generate high-titer lots of sequence-verified, replication-incompetent lentiviral particles pseudotyped with amphotropic, ecotropic, or engineered 10A1 gammaretroviral envelopes—ready for immediate transduction of quiescent hematopoietic stem cells, polarized airway epithelia, or patient-derived organoids without further conditioning.
| Phase | Deliverables |
|---|---|
| Consultation & Cell Panel Matching | Vector map, tropism report, quote |
| Envelope Engineering & QC | Sequence-verified envelope plasmid, endotoxin <0.05 EU/µg |
| Lentivirus Production & Concentration | High-titer supernatant, ddPCR titer, sterility certificate |
| Functional Validation | Transduction efficiency, qPCR integration profile |
| Scale-Up & Archive | Master cell bank, frozen viral aliquots, full batch record |
What We Build for You
Vector Design & Strategy
Your biological target sets the coordinates; we engineer the MLV-pseudotyped lentiviral vector that traverses the cellular entry landscape to reach it. Beginning with structure-guided modeling of the envelope-receptor interface and proceeding through high-density, serum-free production, every pseudotyping variable—receptor-binding loop composition, fusion-peptide pKa, glycan shield density, and post-fusion stalk rigidity—is tuned against the tropism signature of your intended cell type and the kinetic window of your experimental paradigm. Tropism optimization is not a downstream adjustment; it is encoded as a first-principle design constraint from day zero, ensuring that each molecular decision—envelope variant, linker length, and stability mutation—pre-empts transduction inefficiencies and compresses the interval from concept to functional data.
- Envelope Selection Matrix
We start with a short questionnaire (species, target tissue, primary vs. cell line, in vivo or ex vivo). In a short term, you receive a tropism heat-map that ranks MLV-E, MLV-A, 4070A, and any custom envelope against your exact model. Mini-scale MLV-E, MLV-A, 4070A, and custom envelopes produced simultaneously. Transduction efficiency and viability curves generated in your target cell population or primary tissue. You receive raw FCS files plus a concise recommendation report—no reagent purchases on your end. Ready-to-ship envelope plasmids, sequence-verified ORFs codon-optimized for mammalian expression. Optional HA, or FLAG fusions for downstream detection. Full GenBank file and alignment trace accompany every plasmid. Envelope sequence alignment to NCBI reference, endotoxin certificate, and RCL-negative assay summary formatted for IBC/IACUC upload.
- Payload Architecture Options
We modularize every element so you can swap promoters, reporters, or effectors without starting from scratch. Tissue-specific or synthetic promoters: Library of 200+ pre-tested promoters (murine Sca-1, human CD45, synthetic CAG variants, neuron-restricted Syn1, etc.), each promoter is insulated with cHS4 or woodchuck elements to prevent silencing. Strength ranking delivered as an RNA-seq heat-map in your cell type, plus the top-performing promoter already cloned into the final backbone. Multi-cistronic 2A or IRES cassettes: Cleavage efficiencies (P2A, T2A, E2A, F2A) predicted in silico and validated by LC-MS; you receive exact stoichiometry data, pre-built "dual-cassette" backbones (e.g., Cas9-2A-BFP-U6-sgRNA) ready for gene swaps, annotated SnapGene file and Western/LC-MS validation traces included. Inducible or destabilization-domain systems: maximal dynamic range, ddFKBP or ddDHFR destabilization domains with Shield-1 or TMP titration curves generated in your target cells, if induction fails to meet specification, we re-engineer at no additional cost.
Cloning & Sequence Verification
We turn your sequence sketch (or even a hand-drawn napkin) into a fully annotated, sequence-certified lentiviral plasmid that is ready for transfection, packaging, or long-term archiving. Every step is designed to save you bench time, eliminate cloning failures, and give you documentation robust enough for patent filings, journal submissions, or regulatory reviews.
- Seamless Assembly
Design-on-Demand, send a sketch, FASTA, or plain-text description; our bioinformatics team returns an annotated map with optimized junctions and thermodynamic scores. You approve the plan before we start bench work. Golden Gate for multi-fragment inserts or Gibson for large/complex cassettes. Proprietary annealing protocols cut background colonies by > 50 %. All junctions are seamless; no extra amino acids or restriction-site scars. Critical for downstream crystallography, antibody labeling, or fusion-protein work.
- 100 % Sanger Verification – Absolute Sequence Confidence
Every base of the insert, both LTRs, and every junction overlapped at ≥ 4× depth. Color-coded alignment against your reference flags any SNV or indel; if error rate exceeds 0.05 %, we rebuild at no cost. PDF sequence certificate, trace files, and alignment ready for patent filing, core-facility submission, or journal review.
- Codon-Optimization – Maximum Expression, Minimal Risk
Optimize for mouse, human, rat, or CHO; balance CAI, mRNA structure, and tRNA availability. Cryptic splice sites and immunogenic epitopes computationally removed; before/after expression bar charts provided. Optimized ORF already cloned into your chosen lentiviral backbone; no further sub-cloning required.
Production & Purification
We scale and polish your MLV-pseudotyped lentivirus so you can move straight from thaw to transduction—no concentration steps, no endotoxin stripping, and no RCL testing in your own lab.
| Service Element | What You Receive | How It Serves Your Project |
|---|---|---|
| Suspension-adapted packaging + high-density transfection | Serum-free, high-titer supernatant | Consistent yield across any scale; no re-optimization on your side |
| Ultracentrifugation-coupled chromatography | Concentrated stock + endotoxin < 0.1 EU/µg | Ready for direct animal injection or sensitive cell culture |
| RCL-negative certification | Signed analytical report with raw data | Meets institutional and regulatory safety requirements; no in-house safety assay needed |
| Lot-release dossier | Buffer composition, storage guidance, QC certificates | Immediate upload to IBC / IRB / regulatory file |
- Suspension-Adapted Packaging Lines + High-Density Transfection
Serum-free suspension culture eliminates batch-to-batch variability caused by adherent-cell passage drift. Yield on demand: From pilot milliliters to multi-liter runs without re-cloning or re-optimizing ratios. Protein-free medium simplifies downstream purification and reduces host-cell protein carry-over.
- Ultracentrifugation-Coupled Chromatography
Dual-phase endotoxin removal followed by anion-exchange polish delivers endotoxin levels low enough for sensitive in vivo dosing. Concentrated to your requested titer range, aliquoted, and shipped in buffer compatible with direct animal injection or cell-culture supplementation. Raw LAL chromogenic data included—no retest required at your vivarium or core facility.
- RCL-Negative Certification by qPCR
Our orthogonal qPCR assay screens for replication-competent lentivirus at analytical sensitivity exceeding regulatory guidance. Raw Ct values, primer sequences, and positive-control spiking data provided for inclusion in the regulatory submissions. If any lot fails the RCL specification, we repeat production and purification at no charge.
Functional QC Suite – Proof-of-Performance Delivered with Every Lot
Every lentiviral preparation is accompanied by a data package that answers the three questions most critical to downstream success: "How much do I use?", "Where did it land?", and "What else did it touch?" Below is an expanded look at how each assay is run for you, what you receive, and how it accelerates your own research timeline.
- Flow-Cytometry-Based MOI Matrix in Your Cells
Harvest your specified cell line or primary sample and expose it to a ten-point MOI gradient. High-resolution flow cytometry quantifies transduction efficiency and simultaneous viability gating. Raw FCS files are analyzed with standardized templates to eliminate gate-drawing bias. You will receive the interactive dose-response curve (% positive vs. MOI). Recommended working MOI that achieves ≥ 70 % transduction with < 5 % viability loss. All raw FCS files and gating strategy so you can reproduce the analysis in your own core facility if desired.
- ddPCR Integration Profiling ± Clonal Expansion
Droplet digital PCR against the lentiviral WPRE element and a single-copy reference gene delivers absolute copy number per diploid genome. If monoclonality is required, single-cell deposition into 96-well plates is followed by a second ddPCR readout on each clone. You will receive the histogram of integration events across the population, statistical summary (mean, median, range, and 95 % CI), well-by-well copy-number report plus expansion-ready cells.
- In Vivo Biodistribution Pilot (On Request)
Low-dose systemic or local administration in the animal model of your choice. qPCR-based quantification of vector copies in blood, liver, spleen, bone marrow, and any tissue you specify. You will receive the vector copy number per microgram genomic DNA for each tissue, heat-map summary normalized to input dose, raw Ct values and primer sequences for inclusion in IACUC or regulatory dossiers. Demonstrate tissue tropism and absence of off-target accumulation without establishing an in-house animal qPCR pipeline.
What Sets Us Apart?
Envelope Engineering Mastery
Doctoral-level virologists embed directly within the research group, translating cellular-tropism requirements into rationally pseudotyped lentiviral surfaces. Receptor–SU interface energetics, fusion-peptide pKa, and glycan-shield topology are modeled in silico; each variant is benchmarked against off-target entry and innate-sensor activation. Iterative data packages—titers, entry kinetics, and receptor-blocking indices—are delivered on a rolling basis, furnishing principal investigators with quantitative go / no-go metrics without additional personnel.
End-to-End Pseudotyping Pipeline
Whether the starting point is a receptor sequence or a finalized vector backbone, the workflow scales uninterrupted from pilot supernatant to publication-grade particles. Amphotropic, ecotropic, or engineered 10A1 MLV envelopes are expressed in serum-free suspension cultures; clarified harvests are polished by dual-mode membrane chromatography to yield functional titers. A single scientific lead synchronizes envelope cloning, analytical characterization, and functional validation under a unified project map aligned to manuscript or grant milestones.
Analytically Rigorous QC Suite
Each pseudotyped lot is certified against a curated 25-parameter panel: functional titer (qPCR and flow cytometry), envelope incorporation (Western blot densitometry), residual plasmid/host DNA (droplet digital PCR), endotoxin (< 1 EU/mL), and absence of replication-competent lentivirus (qPCR, ≥ 106-fold analytical sensitivity). Primary datasets are pre-formatted as editable methods sections and supplementary tables for direct integration into peer-review submissions.
Tailored Tropism, Modular Design
No fixed catalog constrains design. Every construct is assembled de novo: envelope libraries encompassing amphotropic 4070A, ecotropic, 10A1, or charge-reversal mutants; LTR variants with or without insulator scaffolds; and fluorescent or selectable markers positioned 5′ or 3′ of the payload. Plug-and-play cassettes enable same-day swapping of envelopes or reporters, permitting rapid iteration when tropism barriers or reviewer stipulations arise.
Client Voices

Frequently Asked Questions
Q: What receptor does the MLV envelope engage, and how does this affect tropism?
A: The amphotropic MLV envelope (MLV-A) recognizes the PiT-2 (SLC20A2) phosphate transporter, whereas xenotropic/polytropic variants bind XPR1 (xenotropic and polytropic retrovirus receptor 1). These receptors are highly expressed on human CD34+ hematopoietic stem cells, T-cells, and erythroid progenitors, enabling >3-fold higher transduction of HSCs compared to VSV-G pseudotypes.
Q: Does MLV pseudotyping compromise lentiviral titers or stability?
A: No. Under serum-free suspension conditions, MLV-pseudotyped lentiviral vectors (LV-MLV) achieve functional titers of 1–3 × 108 TU/mL, comparable to VSV-G, and exhibit superior stability in human serum due to reduced complement activation.
Q: How is insertional mutagenesis risk mitigated in MLV-pseudotyped LVs?
A: We employ third-generation self-inactivating (SIN) lentiviral backbones lacking U3 enhancer sequences. Integration site profiling by LAM-PCR shows no significant enrichment near oncogenes, and average proviral copy number remains ≤1 per diploid genome when dosed at MOI 1.
Q: Can the envelope be switched post-production?
A: Yes. The envelope expression plasmid is modular, the plasmid used for production can be replaced in the next production.
References
- Milone, Michael C., and Una O'Doherty. "Clinical use of lentiviral vectors." Leukemia 32.7 (2018): 1529-1541. https://doi.org/10.1038/s41375-018-0106-0.
- Distributed under Open Access license CC BY 4.0, without modification.
