Targeted Nucleases for Genome Editing
Gene-editing Nucleases
The recently developed gene-editing tools can make double-strand breaks to the genome, which further requires repair of these breaks for a successful KI and KO event. The targeted nucleases for gene editing are composed of an engineered target-sequence-binding domain and restriction enzymes. After the programmed nuclease cleaves the target gene to introduce double-stranded breaks (DSBs), the molecular repair proceeds through two different basic mechanisms: homology-directed repair (HDR), where the broken DNA is repaired using a homologous DNA sequence as a template; non-nonhomologous end joining (NHEJ), where the broken ends are rejoined to each other at non-homologous DNA sequence. The HDR repair mechanism, which provides insertion of a template DNA to correct or insert a sequence of choice at the DNA break site, facilitates precise copying of the template at a specific site of the genome and repairs the homologous DNA break. While the NHEJ repair mechanism results in the small insertion or deletion (indels) at the desired place or breaks; hence this mechanism is an efficient way to KO gene function.
Classification of Nucleases
Three major platforms of gene-editing nucleases currently exist for inducing these site-specific DSBs, including transcription activator-like effector nucleases (TALENs), meganucleases and clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease Cas9 (CRISPR/Cas9). These gene-editing nucleases with a tremendous effect on the genetically engineered materials provide powerful tools that improve our ability to understand the physiological processes and their underlying mechanisms for gene therapy.
TALE as a programmable DNA-binding domain originated from plant pathogens Xanthomonas, essentially lead to the engineering of TALENs. TALENs are composed of the tandem repeat of 34-35 amino acids and two amino acids within each module determine the specific DNA base pair recognized. TALENs, are fused to the catalytic domain of the FokI endonuclease as a cleavage domain to cleave their intended DNA target site. Therefore, TALENs have the capability of binds to single specific nucleotide sequences. Besides, TALENs are possibly superior because of its higher specificity, less cytotoxicity, simplified design, and assembly strategies.
The nuclease, which recognizes the genome sequences and makes a double break at a particular site, is called meganucleases. Engineered meganuclease/homing endonuclease is the smallest class of engineered nucleases; they are small and single-chain that make them potentially facilitate gene delivery to the cell. Through a combination of rational design and selection, these engineered/homing endonucleases can be successfully re-engineered to target specific novel loci of the desired genome sequences. Meganuclease technology involves re-engineering the DNA-binding specificity of naturally occurring homing endonucleases. Thus, for particular applications, for example, in gene therapy or biotechnology, engineering a meganuclease could still provide advantages.
Recently, the CRISPR/Cas system, a much simpler and more efficient gene-editing technology, has been developed to the rapid and efficient target DNA sequence. The CRISPR/Cas nucleases as ideal programmable nucleases are derived from a prokaryotic antiviral mechanism to defend against repeat infection. Primarily, the bacterial CRISPR/Cas9 system is composed of two elements: a single guide RNA molecule (sgRNA) that binds a complementary sequence in a target gene and a nuclease protein Cas9 that cuts the target double-stranded DNA. Different from TALENs using DNA-binding proteins to recognize DNA sequence, CRISPR/Cas9 nucleases are attractively adapted for gene editing for the introduction of site-specific DSBs and precise genome modification of any organisms.
In the past few years, engineered nucleases have become more versatile and indispensable and hold significant gene therapy promise. The targeted nucleases as essential tools allow us to modify or express the gene of interest in a site-specific manner with ease-of-use, efficiency, and specificity. Nuclease-based gene editing allows manipulating specific genes in the host genome for various applications, including genome and basic metabolic research, disease diagnosis, genetic disorders therapy, thus enabling the investigation of disease pathophysiology and facilitating diverse therapeutic development.
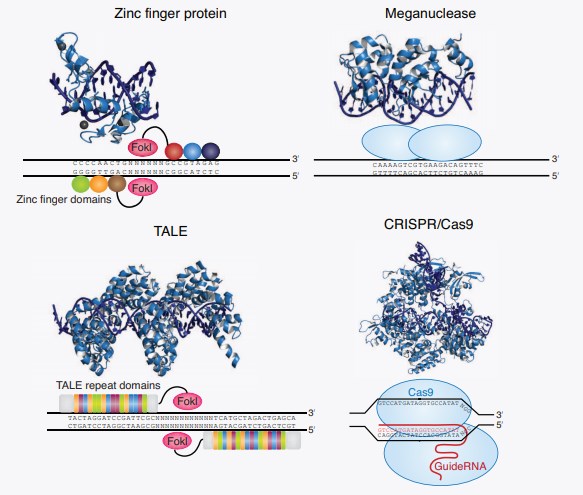 Fig.1 Targeted nucleases for genome editing.1
Fig.1 Targeted nucleases for genome editing.1
Reference
- Maeder, M. L.; Gersbach, C. A. (2016). Genome-editing technologies for gene and cell therapy. Molecular Therapy. 24(3): 430-446. Distributed under Open Access license CC BY 4.0, without modification.
