siRNA in Epigenetic Regulation-The Hidden Layer
Introduction of siRNA in Epigenetic Regulation
Small interfering RNAs (siRNAs) function as key elements in post-transcriptional gene silencing by operating through the RNA interference (RNAi) pathway. In addition to their role in mRNA degradation siRNAs have critical functions within epigenetic regulation mechanisms. Epigenetic changes such as DNA methylation and histone modifications control gene expression while maintaining the original DNA sequence. siRNAs guide chromatin-modifying enzymes to exact locations on the genome to provide additional regulation of gene expression through epigenetic mechanisms.
siRNA's Secret Role Beyond Gene Silencing
- siRNA's Dual Function: mRNA Degradation + Epigenetic Modulation
Through post-transcriptional gene silencing and epigenetic regulation siRNAs serve as flexible molecules. Through the RNA-induced silencing complex (RISC) siRNAs achieve their dual function by targeting specific mRNAs for degradation while also activating chromatin-modifying enzymes at precise genomic sites. SiRNAs can control gene expression through both their interactions with mRNA and their capacity to modify epigenetic structures.
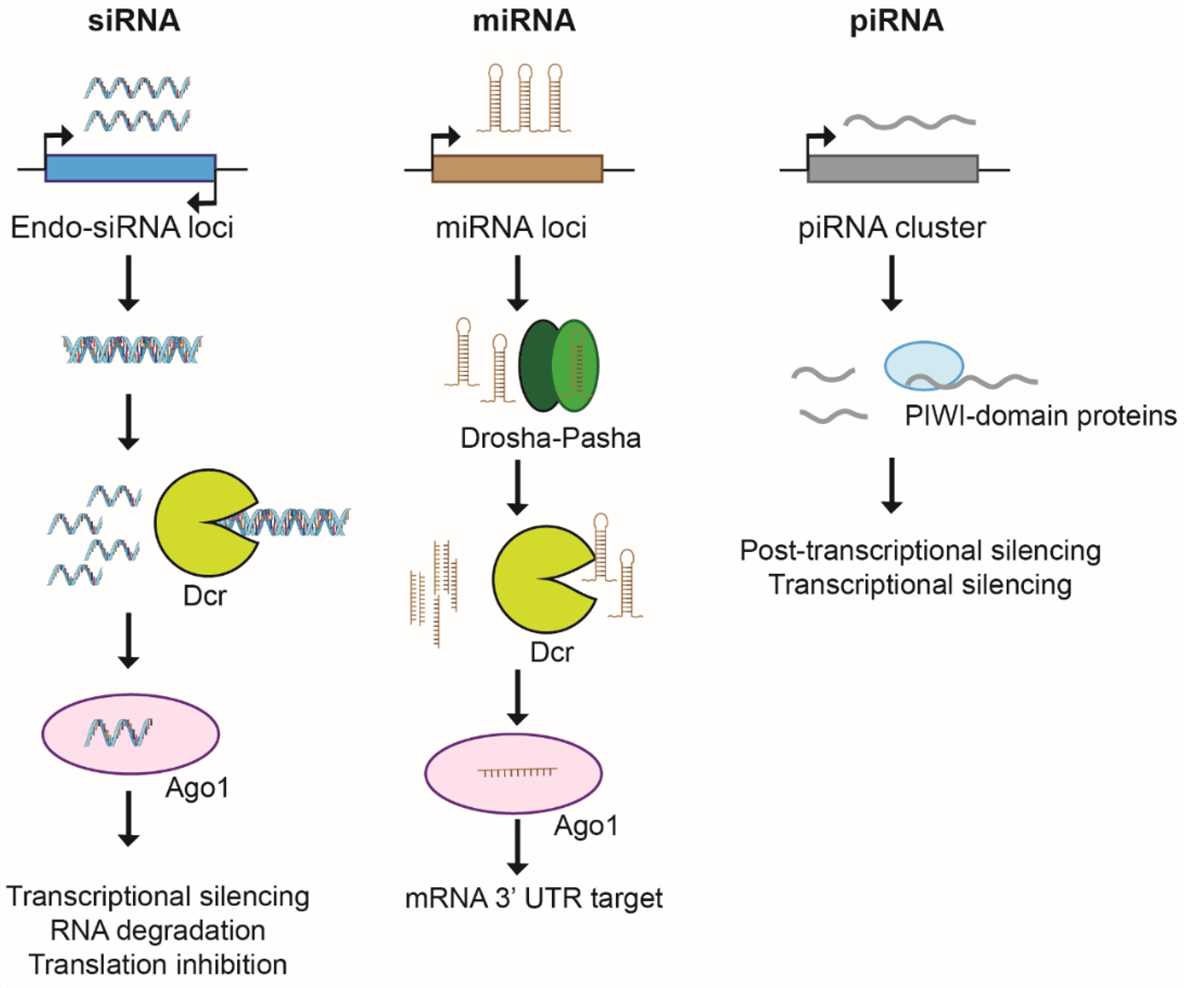 Fig.1 RNA interference pathways1,6.
Fig.1 RNA interference pathways1,6.
- siRNA Adds a Regulatory "Software Layer"
Stable gene expression patterns throughout development and environmental changes depend on essential epigenetic modifications. siRNAs serve as a modifiable regulatory mechanism that affects gene expression through changes in chromatin architecture and accessibility. This "software layer" enables cells to modify their gene expression patterns without making permanent DNA sequence alterations. siRNAs direct DNA methyltransferases to particular genomic sites where DNA methylation then causes transcriptional repression.
- Historical Perspective
The discovery of siRNAs' function in epigenetic regulation began when scientists observed RNA-directed DNA methylation in plant systems. The study of Arabidopsis thaliana demonstrated the process by which siRNAs direct DNA methyltransferases to precise genome regions resulting in permanent transcriptional silencing. Multiple species such as fission yeast and mammals demonstrated that siRNAs serve as vital components for epigenetic regulation.
The Epigenetic Toolbox: Where siRNA Fits In
- DNA Methylation: siRNA Guiding Methyltransferases to Specific Loci
siRNAs lead DNA methyltransferases to specific genomic sites which causes CpG island methylation to occur followed by the suppression of transcription. The process leads to heterochromatin formation while simultaneously silencing transposable elements. The siRNAs in S. pombe target the Clr4 DNA methyltransferase to centromeric repeats to enable correct chromosome segregation.
- Histone Modifications: siRNA Recruiting Chromatin Remodelers
siRNAs initiate the recruitment of chromatin-remodeling complexes which leads to histone modifications. In Arabidopsis siRNAs target histone methyltransferases for histone H3K9 methylation to trigger heterochromatin formation. Genome stability is preserved through the process that silences repetitive sequences and blocks unsuitable gene activation.
- Nuclear Architecture: siRNA Influencing 3D Genome Organization
While siRNAs modify DNA and histones directly they also play a role in organizing the genome's three-dimensional structure. siRNAs recruit chromatin-modifying complexes to change genomic regions' spatial arrangement which results in the movement of regulatory elements closer together or further apart. Spatial regulation produces significant impacts on both gene expression patterns and overall genome function.
siRNA-Directed DNA Methylation (RdDM) in Plants
- Model system: How Arabidopsis revealed this pathway
The RdDM pathway in plants utilizes siRNAs to direct the new methylation of DNA sequences. Researchers first identified and thoroughly investigated this pathway in the model plant Arabidopsis thaliana. Arabidopsis needs RdDM to silence transposable elements and control gene expression. The process includes RNA polymerases Pol IV and Pol V which function together with various proteins to create and sustain DNA methylation patterns.
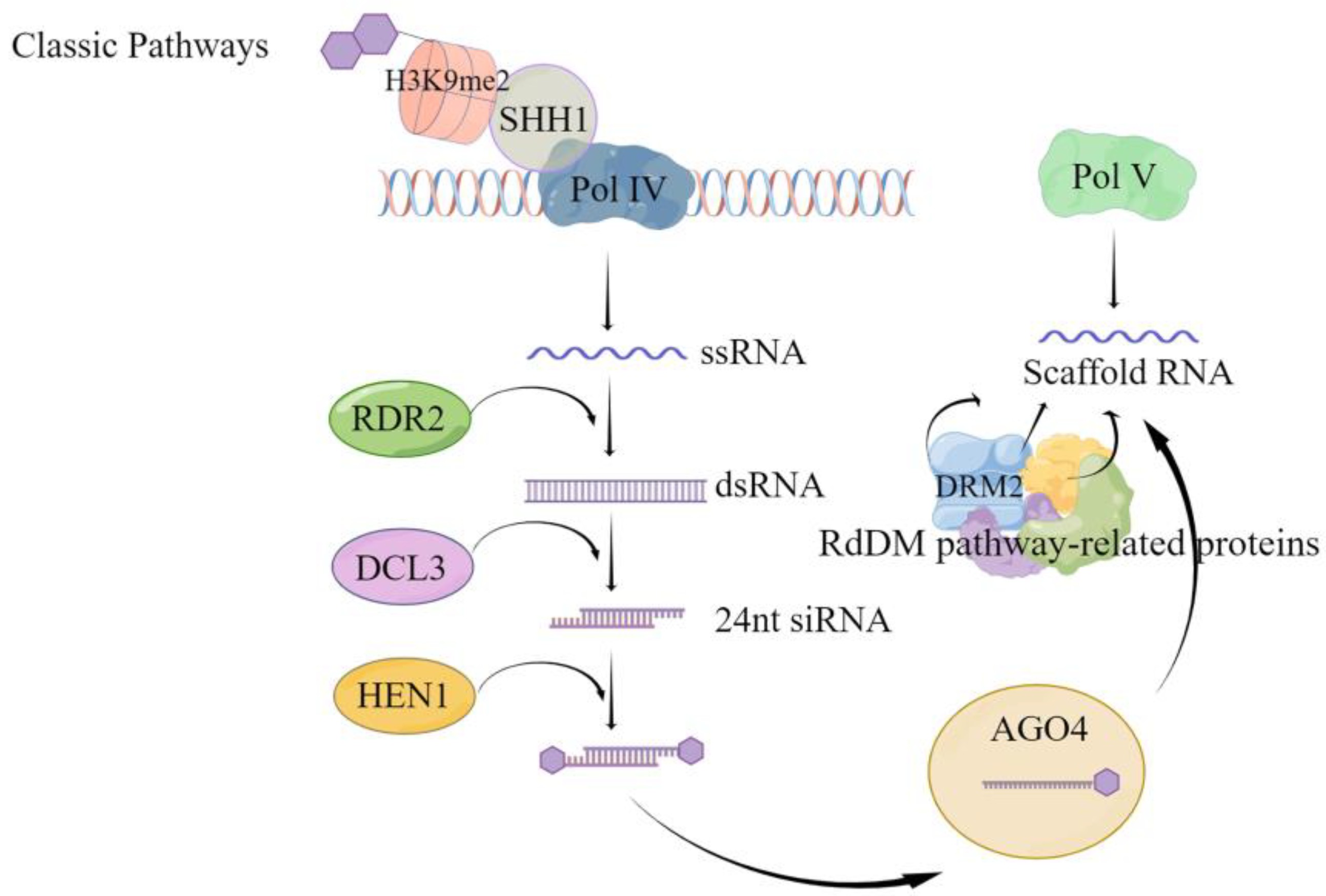 Fig.2 Classical RdDM pathway-mediated de novo methylation pattern2,6.
Fig.2 Classical RdDM pathway-mediated de novo methylation pattern2,6.
- Agricultural applications: Epigenetic engineering of crop
Scientists can utilize RdDM to selectively turn off targeted genes or transposable elements within crop plants. Engineered siRNAs that focus on suppressing undesirable traits or harmful TEs enable improved crop stability and performance. RdDM technology has been applied to regulate flowering time genes which enables enhanced control over crop maturation periods. Scientists design siRNAs to attack viral or pathogen-derived sequences which creates RNA-based immunity mechanisms. Multiple crop species have gained resistance to viruses and other pathogens through this technique. The RdDM method enables the introduction of epigenetic changes that remain stable through multiple generations. Through this method scientists can produce crop varieties with beneficial characteristics without altering their DNA sequences genetically. Agricultural scientists utilize RdDM technology to turn off genes that reduce crop yield or reduce stress tolerance.
siRNA's Epigenetic Roles in Animals
- Contrasts with Plants: More Limited but Significant Effects
The importance of siRNA in epigenetic control in mammals exists but remains more restricted than in plant cells. Plants extensively utilize siRNA for RdDM to silence TEs and regulate gene expression while mammals primarily depend on piRNAs for these functions. Despite their reduced role in mammals compared to plants siRNAs remain essential for highly specific biological processes including germline development and gene imprinting.
- Repeat Element Control: siRNA Silencing Transposons Epigenetically
siRNAs in mammals function as molecular tools to epigenetically silence transposable elements which act as mobile genetic elements that pose a threat to genome stability. siRNAs direct chromatin-modifying enzymes toward TEs which results in transcriptional silencing similar to the process observed in plants. The piRNA pathway which shares similarity with siRNA pathways plays a critical role in the suppression of TEs within mouse germline cells. The piRNA pathway silences transposable elements through transcript degradation and histone modification and DNA methylation to block their movement. The function of this mechanism serves as a vital element for preserving genomic stability throughout gametogenesis.
- Imprinting Maintenance: Protecting Methylation at Imprinted Loci
Imprinting represents an epigenetic process that leads to the expression of specific genes depending on whether they originate from the mother or the father. siRNAs along with their related small RNAs function to preserve methylation patterns at imprinted sites in mammalian genomes. The piRNA pathway functions as a necessary mechanism for establishing new DNA methylation patterns at the imprinted rasgfr1 locus within male mouse germline cells. piRNAs target specific genomic loci which triggers the recruitment of DNA methyltransferases to create essential methylation patterns for imprinting. Other imprinted loci may be maintained through similar mechanisms which demonstrates the essential role of small RNAs in controlling epigenetic gene expression.
Transgenerational Epigenetics: siRNA's Inherited Memory
- Germline Transmission: siRNA Carrying Epigenetic Information
siRNAs serve as essential mediators for transferring epigenetic information across multiple generations according to transgenerational epigenetics research. Researchers have extensively studied how siRNAs from the germline are transmitted to future generations of Caenorhabditis elegans, resulting in enduring gene silencing effects. The epigenetic information carried by siRNAs remains effective across generations because these molecules can migrate between tissues and replicate through RdRP activity.
- How Stress-Induced siRNA Changes Are Inherited
The presence of environmental stressors triggers alterations in siRNA populations which future generations inherit. Specific siRNAs that mediate adaptive responses are produced in C. elegans when it is exposed to pathogens or starvation. These siRNAs transfer to offspring and equip them with improved defense against comparable stressors. Multiple studies have shown that stress responses can be passed down through generations due to the role of siRNAs which connect environmental experiences to inherited phenotypes.
- Case Study: C. elegans and Multigenerational RNAi
The short generation time and capacity to cultivate vast numbers of genetically identical individuals make C. elegans an optimal model for transgenerational epigenetic inheritance studies. The inheritance of RNAi phenotypes serves as a well-known example in scientific literature. Upon exposure to double-stranded RNA (dsRNA) targeting particular genes C. elegans exhibits gene silencing effects that extend across several generations. RdRP facilitates this inheritance process through the production of secondary siRNAs that use target mRNAs as templates for their generation. Secondary siRNAs move into the germline where they trigger gene silencing in subsequent generations.
Cancer Epigenetics: siRNA as a Corrective Force
- Reversing Hypermethylation: siRNA Targeting DNA Methyltransferases
Cancers frequently show epigenetic alterations through tumor suppressor gene promoter hypermethylation which silences these genes. Essential enzymes known as DNA methyltransferases (DNMTs) support this biological process. Research uses siRNA to downregulate DNMTs which triggers demethylation and reactivation of silenced tumor suppressor genes. Studies show that targeting DNMT1 with siRNA leads to demethylation and activation of tumor suppressor genes RASSF1A and APC in lung cancer cells. This method can normalize gene expression profiles while preventing cancer cell growth.
- Oncogene Control: Dual Epigenetic + Transcriptional Silencing
The therapeutic use of siRNA against oncogenes combines epigenetic and transcriptional silencing processes to interfere with gene functionality. Oncogene mRNAs become targets for siRNA which causes direct reduction of their expression. Epigenetic modifications at oncogene sites through siRNA treatment intensify the gene silencing impact. siRNA targeting DNMTs leads to tumor suppressor gene demethylation and oncogene expression reduction by initiating RNAi processes. The dual mechanism creates a complete cancer therapy solution by targeting the genetic and epigenetic factors of tumor development.
- Therapeutic Potential: More Durable than Standard RNAi
siRNA-mediated epigenetic regulation presents multiple benefits in cancer treatment which surpass those of traditional RNAi methods. Standard RNAi functions mainly through the breakdown of mRNA, whereas siRNA-triggered epigenetic alterations result in more permanent suppression of gene expression. siRNA treatment against DNMT1 decreases the enzyme's expression levels and establishes permanent DNA methylation modifications that remain even in the absence of siRNA treatment. SiRNA demonstrates long-lasting activity which positions it as a powerful option for cancer treatment because it maintains suppression of cancer-driving pathways while potentially bypassing resistance mechanisms.
Viral Epigenomes: siRNA's Defense and Subversion
Antiviral methylation processes directed by siRNA make it a powerful defense mechanism that host organisms use to protect themselves from viral infections. RNAi serves as an essential antiviral defense system in plants through the activation by dsRNA molecules that form during viral replication. Dicer-like enzymes convert dsRNAs into virus-derived siRNAs which enable the RISC to destroy viral RNA and stop viral replication. The host RdDM pathway triggers DNA methylation to silence viral genomes. The RdDM pathway in plant cells targets viral DNA when infected with DNA viruses and induces methylation followed by transcriptional silencing. During viral latency, siRNA mechanisms alongside their related pathways are essential for keeping proviral DNA in a silenced state. The epigenetic machinery of the host targets proviral DNA in HIV through histone modifications and DNA methylation to achieve silencing. HIV has developed methods to prevent silencing by integrating into transcriptionally active areas of the host genome which prevents epigenetic repression. Herpesviruses establish latent infections by making use of the host's epigenetic mechanisms to keep their viral genomes silenced until they reactivate. A critical balance exists between host defense mechanisms and viral countermeasures which enables the establishment and maintenance of viral latency.
Future Horizons: The Unexplored Epigenetic Landscape
- Undiscovered Pathways: Likely More siRNA-Epigenome Connections
As research in epigenetics advances quickly, scientists expect to find numerous unidentified pathways connecting siRNA with the epigenome. Research from recent studies demonstrates that noncoding RNAs such as siRNAs significantly regulate gene expression by modifying epigenetic patterns. siRNAs direct chromatin-modifying enzymes to particular genomic sites which results in histone modifications together with DNA methylation. The current research indicates that siRNAs perform wider functions in constructing the epigenetic architecture than existing scientific knowledge suggests.
- Aging Research
The close relationship between epigenetic modifications and aging has prompted recent studies into siRNA and other epigenetic tools as methods to reverse cellular aging. Scientific studies demonstrate a connection between RNA modifications like m5C (5-methylcytosine) and cellular senescence. Recent research demonstrates that applying Yamanaka factors to mature cells can revert them to a pluripotent state and reverse several age-specific cellular changes. The research shows that siRNA along with epigenetic interventions may enable scientists to restore youthful cellular states by reversing cellular aging.
- Consciousness Links: Extreme Epigenetic Plasticity in Neurons
Neurons demonstrate sophisticated epigenetic plasticity which plays a critical role in learning processes and cognitive abilities. Modern research demonstrates how DNA methylation and histone modifications as epigenetic modifications significantly impact both neuronal function and plasticity. Researchers have yet to fully establish the direct connection between siRNA and neuronal consciousness but siRNA and other small RNAs are known to affect gene expression and epigenetic conditions within neurons. Researchers propose that siRNA might serve as a tool to regulate neuronal activity and adaptability because it offers potential therapeutic benefits for neurological diseases and improved cognitive abilities.
References
- Iracane, Elise, Samuel Vega-Estévez, and Alessia Buscaino. "On and off: epigenetic regulation of C. albicans morphological switches." Pathogens 10.11 (2021): 1463. https://doi.org/10.3390/pathogens10111463.
- Fan, Youfang, et al. "Recent advances in studies of genomic DNA methylation and its involvement in regulating drought stress response in crops." Plants 13.10 (2024): 1400. https://doi.org/10.3390/plants13101400.
- Grishok, Alla. "Small RNAs worm up transgenerational epigenetics research." DNA 1.2 (2021): 37-48. https://doi.org/10.3390/dna1020005.
- Lai, Qi, et al. "The loss-of-function of DNA methyltransferase 1 by siRNA impairs the growth of non-small cell lung cancer with alleviated side effects via reactivation of RASSF1A and APC in vitro and vivo." Oncotarget 8.35 (2017): 59301. https://doi.org/10.18632/oncotarget.19573.
- Saul, Dominik, and Robyn Laura Kosinsky. "Epigenetics of aging and aging-associated diseases." International journal of molecular sciences 22.1 (2021): 401. https://doi.org/10.3390/ijms22010401.
- Distributed under Open Access license CC BY 4.0, without modification.
