Viral Warfare-siRNA's Broad-Spectrum Antiviral Campaign
Introduction of siRNA in Viral Warfare
Scientists use small interfering RNA (siRNA) to fight viral infections because this method activates the cell's natural RNA interference (RNAi) mechanism to stop gene expression. RNAi operates as a universal cellular mechanism that utilizes small RNA molecules to destroy target mRNA and prevent gene expression. Scientists first discovered this biological process in the nematode Caenorhabditis elegans before detecting its existence across multiple species including humans. siRNA molecules that measure between 21 and 25 nucleotides function by pairing with viral RNA sequences to precisely target and degrade viral genomes or transcripts. Most conventional antiviral treatments depend on antibodies and interferons which present major disadvantages. Antibodies provide precise targeting capabilities but only work against viruses outside cells and struggle with viruses that mutate quickly. Interferons create a wide-ranging antiviral response within cells yet they present substantial side effects and lack universal effectiveness against all viruses. siRNA provides a versatile and precise method for antiviral treatment. Rapid design capabilities enable siRNA to target conserved viral genome regions which results in effectiveness against various viral strains. siRNA functions as a versatile antiviral tool by enabling targeting of essential viral replication elements and crucial host factors.
Introduction: The Eternal Arms Race Against Viruses
- Potential of Viral Threats
The world's health system confronts major dangers from viral diseases because new and recurring viruses trigger serious epidemic outbreaks. The emergence of COVID-19 through SARS-CoV-2 infections proves the necessity of creating potent antiviral treatments. Ebola and HIV alongside influenza viruses remain major global health threats due to their persistent impact on worldwide morbidity and mortality rates. The rapid mutation rate of viruses creates drug-resistant variants that complicate treatment development. Antiviral treatments such as nucleoside analogs and protease inhibitors show different effectiveness levels among viral strains due to mutations that create resistance to these treatments. The emergence of drug resistance creates significant challenges for treating both HIV and hepatitis C virus infections. Existing antiviral medications face limitations which demand the creation of adaptable treatment approaches to manage rapidly mutating viruses and pathogens.
- siRNA stands out as an intelligent tool for Antiviral Therapy
siRNA-based treatments provide a hopeful remedy to address the problems created by viral diseases. siRNA therapies allow quick development to attack conserved viral genome segments which enables treatment of various strains while minimizing resistance development. Therapeutic effectiveness improves through dual-targeting when siRNA targets essential host replication factors. The studies performed on laboratory animals demonstrated significant antiviral effects when siRNA targeted the RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2. SiRNA technology functions as a sophisticated antiviral defense system due to its adaptable precision which allows for diverse treatment possibilities.
The molecular mechanism of siRNA
- Step-by-step mechanism
During replication within a host cell, viral infection often results in the generation of double-stranded RNA (dsRNA) intermediates. The host cell's innate immune system recognizes dsRNA molecules as external harmful threats. Dicer functions as an essential enzyme within the RNAi pathway where it identifies dsRNA molecules to produce short siRNA fragments that contain 21 to 25 nucleotides. Dicer creates siRNA fragments which have sequences that specifically match viral RNA strands. The complementary base pairing between siRNA and target viral RNA allows the siRNA to specifically bind to its target. The siRNA molecules consist of two strands which are known as the guide strand and the passenger strand. The guide strand acts as the key functional unit that targets the RNA-induced silencing complex (RISC) toward the viral RNA. The formation of siRNA fragments leads to their integration into the RISC complex. The Argonaute-containing RISC complex separates the siRNA duplex by unwinding it and discards the passenger strand while keeping the guide strand. The guide strand functions to guide the RISC complex to sequences of viral RNA that complement its own structure. The Argonaute protein inside the RISC complex binds to viral RNA and cleaves it which results in degradation and stops viral replication.
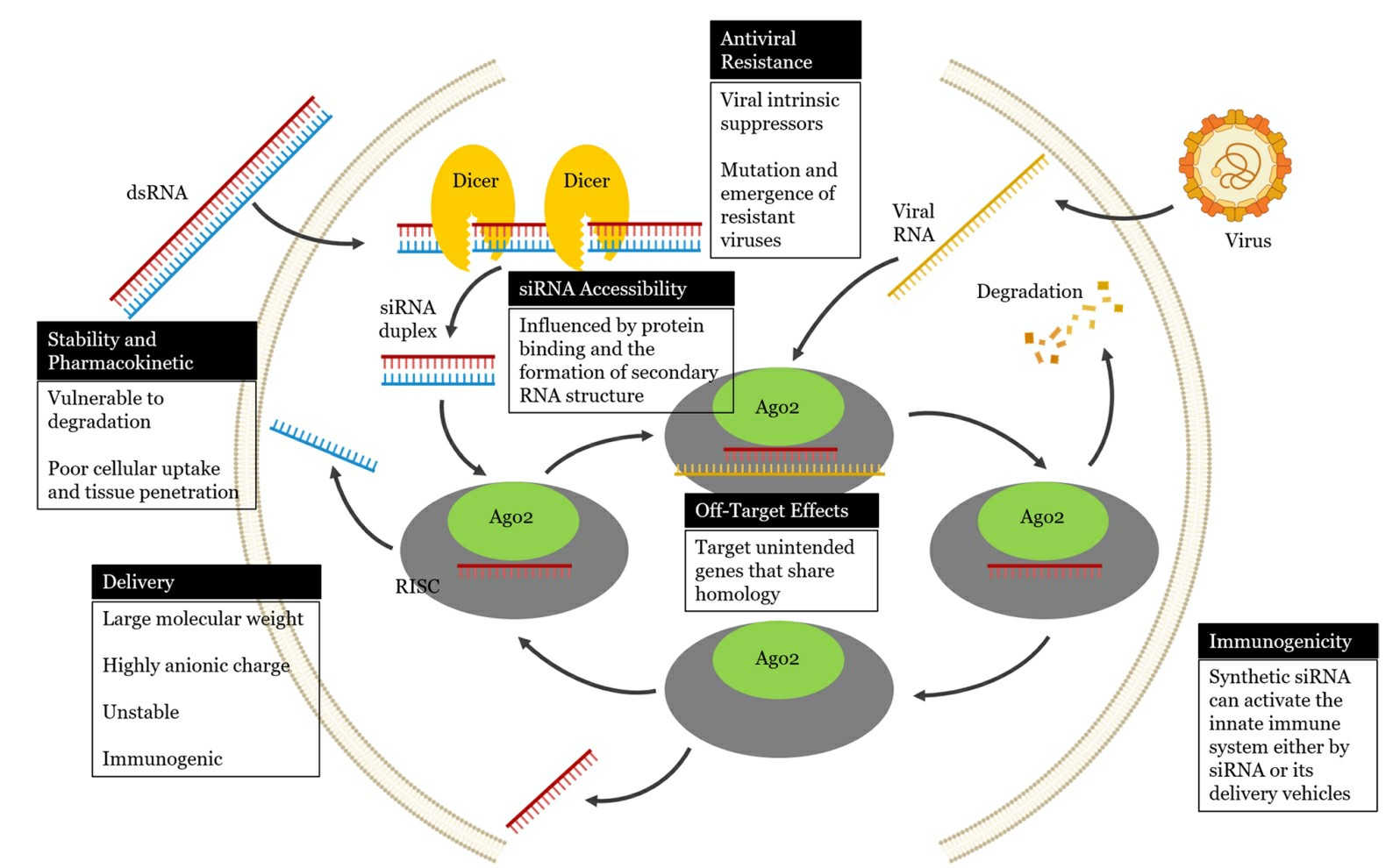 Fig.1 Schematic illustrations showing challenges to small interfering RNA (siRNA) applications against viral infections1,6.
Fig.1 Schematic illustrations showing challenges to small interfering RNA (siRNA) applications against viral infections1,6.
- Visual analogy: Scissors cutting viral blueprints
The siRNA mechanism functions like molecular scissors known as the RISC complex that precisely targets and deactivates viral RNA blueprints. Upon detecting viral RNA the host cell employs Dicer enzyme to generate siRNA fragments which function as "molecular guides" for directed cutting by the molecular scissors. These molecular guides match specific viral RNA sequences to ensure that only viral RNA gets cut by the scissors without damaging host genetic material. After loading the siRNA fragments into the RISC complex it proceeds to search throughout the cell for the viral RNA. When the RISC complex identifies a viral RNA sequence that aligns with the siRNA guide it will bind to it before the Argonaute protein cleaves the viral RNA thereby destroying the viral blueprint. The virus cannot replicate or spread to other cells because this precise cutting action serves as a specific and efficient defense mechanism against viral infections.
Broad-Spectrum Potential of siRNA for Multiple Viruses
Advantages over traditional antivirals
- Targets Conserved Viral Genes
siRNA-based therapies surpass traditional antiviral drugs because they attack stable areas within viral genomes. The conserved regions of viral genomes show low mutation tolerance which prevents viruses from easily developing resistance. Researchers have created siRNAs that target the conserved sections of SARS-CoV-2's RNA genome including the ORF1a/b region because this region produces critical non-structural proteins needed for viral replication. The method enables siRNA to stay effective against stable targets despite viral mutations occurring elsewhere.
- Adaptable to New Strains
siRNA stands out because it easily adapts to combat new viral strains. siRNA offers a rapid design and synthesis solution for emerging viral strains unlike traditional antivirals which need extensive development and testing periods. The rapid development of siRNAs as a solution to target the SARS-CoV-2 genome demonstrated their functionality as an immediate response approach during the initial phase of the COVID-19 pandemic. Therapeutics must keep up with viral evolution to effectively treat fast-mutating diseases such as influenza and HIV.
Laboratory studies
- Influenza
Research in controlled lab environments shows siRNA successfully prevents influenza virus replication. siRNAs directed against influenza virus nucleoprotein (NP) and polymerase basic protein 1 (PB1) demonstrated substantial effectiveness in diminishing viral levels within infected cells. The intranasal siRNA treatment in mice resulted in significant decreases in influenza virus infection which highlights the dual potential of siRNA for prevention and therapy.
- HIV
Researchers have employed siRNA against HIV viral genes like gag and nef and essential host factors required for viral replication. Research demonstrates that siRNA aimed at the HIV-1 gag gene achieves significant reductions in viral load within infected cells. Scientists used siRNA to block the expression of host proteins including CD4 which is the main receptor HIV uses to enter cells, thereby stopping viral infection.
- SARS-CoV-2 (COVID-19)
The COVID-19 pandemic demonstrated siRNA's potential as an antiviral treatment. Scientific research has confirmed successful siRNA targeting of the SARS-CoV-2 genome in various studies. siRNA directed against the viral RdRP together with the spike protein demonstrated substantial antiviral effects when tested in cell cultures. Researchers found that siV1 siRNA aimed at SARS-CoV-2 leader protein halted viral replication in cell lines by over 99% without producing toxic effects. The results show that siRNA could serve as an effective weapon against SARS-CoV-2 and its multiple variants.
Challenges of siRNA treatments for viral infections
- Vulnerability of siRNAs to degradation
Antiviral siRNA therapy faces primary extracellular barriers due to fast systemic elimination that exposes siRNAs to degradation by nucleases. RNAs that lack modifications cannot resist degradation by extracellular nucleases and their negative charge prevents them from passing through the hydrophobic cytoplasmic membrane which limits their stability and pharmacokinetic profile. Due to these barriers siRNAs cannot efficiently penetrate cells or access tissues which diminishes their therapeutic potential. Ribose in the RNA backbone creates susceptibility to degradation from serum nucleases that attack phosphodiester bonds. After systemic administration therapeutic siRNA fails to accumulate intact in target tissues because of this degradation process.
- The ability to penetrate cell membranes
Naked and unmodified siRNAs fail to cross the cell membrane because both the siRNA molecules and the membrane carry negative charges. SiRNAs that remain outside cells in the extracellular space face quick degradation by RNases and phosphatases because they are not absorbed by the cells. After penetrating the cell membrane siRNAs need to avoid endosomal confinement before reaching RNAi machinery. The challenge of endosomal trapping stands as a major hurdle to RNAi-based therapy and multiple outstanding reviews have explored this topic in depth. When exogenous siRNA enters cells, they first become encapsulated within endosomes while the siRNA is then released into the cytoplasm as it escapes from these endosomal compartments. The siRNA delivery system gains cellular entry through endocytosis which forms endocytotic vesicles known as endosomes that become acidified by the ATPase proton pump located in the endosomal membrane. The siRNA moves into lysosomes which function as acidified organelles that lead to siRNA degradation. The delivery system design must ensure protection of siRNA from acidification and degradation once it enters the cell through endocytotic vesicles.
- Delivery hurdles of siRNA
siRNA therapeutic development for human use faces substantial obstacles in antiviral siRNA delivery because of siRNA's complex pharmacological characteristics. siRNAs demonstrate a substantial molecular mass at around 13 kDa and feature multiple phosphate groups that create a high anionic charge with 38 to 50 such groups. The molecular weight and high anionic charge of siRNAs create challenges for their passage through cell membranes. Unmodified siRNAs demonstrate poor stability in blood circulation and trigger immune reactions by activating Toll-like receptors (TLRs). When delivered via intravenous injection siRNAs need to cross the vascular endothelial barrier and diffuse through the extracellular matrix before avoiding kidney filtration and preventing uptake by non-target cells. siRNAs need to protect themselves from nuclease digestion during their journey to remain effective.
- Immune responses induced by siRNA
Researchers found that synthetic siRNA duplexes trigger immune responses which result in elevated production of inflammatory cytokines and type I interferon. The immune responses triggered by siRNA showed significant activity after systemic delivery. The innate immune system activation by siRNAs occurs through multiple mechanisms that include recognition by cytoplasmic and endosomal receptors such as TLRs. Torre et al. In their research Torre et al. found that the siRNA they used triggered TLR3 activation. The activation process resulted in both β-interferon synthesis and caspase activity initiation. Antibodies targeting TLR3 blocked siRNA activation showing that siRNA bound to TLR3 ectodomain binding sites and caused receptor dimerization. The direct delivery of siRNA into cells reduces the chance of triggering an innate immune interferon response. The approach avoids activating Dicer pathways and blocks protein production when long dsRNA pieces exceeding 30 nucleotides bind to cellular RNA receptors.
Strategies to improve the applicability of siRNA
- Methods to enhance siRNA stability and prevent its breakdown
The stability of siRNA can be enhanced by using various 2' sugar modifications such as fluorine substitutions which improve resistance to endonucleases when included in the delivery system. Modifications to the sugar–phosphate backbone of siRNA can include the addition of 2'-fluoro and 4'-thio groups as well as the incorporation of locked nucleic acids while inducing phosphorothioation and methyl phosphonation to extend siRNA stability and half-life in serum. The study showed that 2'-fluoro-modified antiviral siRNA swarms demonstrated superior antiviral activity against herpes simplex virus 1 in human corneal epithelial cells. Studies showed that antiviral siRNA swarms with 2'-fluoro modifications demonstrated more significant impact compared to standard antiviral siRNA swarms. Researchers demonstrated that siRNA activity rose while its stability improved in serum-containing environments when 2'-deoxy-2'-fluoro-beta-d-arabinonucleotide units were incorporated into siRNA duplexes. Researchers use phosphorothioate backbone linkages at RNA strands' 3'-ends to protect siRNA from enzymatic degradation by reducing exonuclease activity. According to scientific research the inclusion of glycol nucleic acid nucleotide or dinucleotide within oligonucleotides enhanced their resistance to degradation by 3'-exonuclease.
- Delivery systems increase the effectiveness of antiviral siRNA treatments
The delivery systems of antiviral siRNAs fall into two main categories which are vectors and nanoparticles. Expression cassettes known as plasmid and viral vectors deliver sustained silencing effects. Adenovirus, adeno-associated virus and lentivirus stand out as popular viral vectors for delivering siRNA both in cell cultures and living organisms despite plasmids proving less effective at delivery. Successful delivery of therapeutic agents requires that lipid-based systems and polymer-based systems both possess essential chemical properties. SiRNA delivery vehicles based on polymers provide structural versatility and protection from nucleases. Cationic materials maintain stable interactions with well-defined polymer structures which facilitate cross-linking and show potential for efficient siRNA delivery. The use of cationic lipids in liposomes allows them to counteract the negative charges that siRNAs possess. Stable nucleic acid lipid particles strengthen siRNA protection while improving its transport capabilities. Stable nucleic acid lipid particles enabled researchers to deliver siRNA effectively to combat hepatitis B virus in mice. Effective delivery systems feature components including cationic/ionizable groups along with functional linkers and lipid tails. Creating direct conjugates of ligands or polymers to siRNA generates a delivery system that consists of one component with a specific predetermined composition.
Future Frontiers of siRNA: Beyond Antivirals
- Combating Antibiotic-Resistant Bacteria
siRNA demonstrates promise as a tool to confront the increasing threat posed by antibiotic-resistant bacteria. siRNA targeting essential bacterial genes disrupts important metabolic and replication pathways which increases bacterial vulnerability to antibiotics or enables siRNA to function independently as an antibacterial agent. siRNA molecules that target bacterial ribosomal RNA (rRNA) or essential virulence factors have proven effective at killing bacteria in preclinical studies. The described method shows promise for managing infections by multidrug-resistant bacteria which cannot be treated by standard antibiotics.
- Cancer Therapy: Silencing Oncogenes
siRNA represents a potent therapeutic strategy in cancer treatment because it enables specific oncogene silencing and improves treatment outcomes. The use of siRNA against the KRAS oncogene demonstrated substantial antitumor effects in preclinical pancreatic cancer models. Researchers have combined siRNA targeting Bcl-2 and Mcl-1 antiapoptotic proteins with chemotherapeutic agents to improve apoptosis rates and overcome cancer cell drug resistance. Research demonstrates how siRNA technology can target oncogenes for selective silencing to achieve better results in cancer treatment.
- Synthetic Biology: Programmable RNAi Networks
The field of synthetic biology utilizes siRNA to develop programmable RNA interference networks that permit precise engineering of gene expression regulation. Engineers can design these RNAi networks to react to distinct cellular signals or environmental triggers which enables them to perform targeted gene silencing. Scientists developed synthetic RNAi circuits that target specific mRNAs for degradation through inducible promoters which results in conditional gene knockdown. Gene therapy, tissue engineering and personalized medicine stand to benefit significantly from this technique.
References
- Kang, Hara, et al. "Small interfering RNA (siRNA)-based therapeutic applications against viruses: principles, potential, and challenges." Journal of Biomedical Science 30.1 (2023): 88. https://doi.org/10.1186/s12929-023-00981-9.
- Mehta, Aditi, Thomas Michler, and Olivia M. Merkel. "siRNA therapeutics against respiratory viral infections—What have we learned for potential COVID‐19 therapies?." Advanced healthcare materials 10.7 (2021): 2001650. https://doi.org/10.1002/adhm.202001650.
- Motamedi, Hamid, et al. "Principle, application and challenges of development siRNA-based therapeutics against bacterial and viral infections: a comprehensive review." Frontiers in Microbiology 15 (2024): 1393646. https://doi.org/10.3389/fmicb.2024.1393646.
- Hu, Bo, et al. "Therapeutic siRNA: state of the art." Signal transduction and targeted therapy 5.1 (2020): 101. https://doi.org/10.1038/s41392-020-0207-x.
- Zhang, Yuan, et al. "Nanoparticle delivery platforms for RNAi therapeutics targeting COVID-19 disease in the respiratory tract." International journal of molecular sciences 23.5 (2022): 2408. https://doi.org/10.3390/ijms23052408.
- Distributed under Open Access license CC BY 4.0, without modification.
