siRNA in Developmental Biology-A Blueprint for Growth
siRNA gene silencing: siRNA as a Master Regulator of Development
The RNA interference (RNAi) pathway employs siRNA as a critical element for steering embryonic development. The RNAi pathway works through the process of mRNA molecule degradation to control gene expression. The Dicer enzyme breaks down double-stranded RNA (dsRNA) into siRNA fragments that range from 21 to 25 nucleotides in length. The RNA-induced silencing complex (RISC) targets mRNA molecules for destruction through guidance from siRNAs resulting in specific gene silencing. In the early stages of embryonic development siRNA fulfills fundamental biological roles through cell division processes and differentiation along with morphogenesis. Studies demonstrate that siRNA controls the asymmetric cell division of Caenorhabditis elegans which plays a vital role in the organism's developmental process and tissue formation. siRNA regulation controls embryonic development stages while also determining cell identity. Studying siRNA throughout developmental stages yields important findings which advance regenerative medicine techniques. Research into siRNA's regulatory mechanism during embryogenesis enables scientists to create precise treatments for tissue repair and regeneration. Researchers utilize siRNA to control key genes that drive stem cell differentiation which leads to damaged tissue regeneration.
siRNA in Early Embryogenesis: The First Genetic Decisions
- Zygote to Blastocyst: siRNA's Role in the First Cell Divisions
The siRNA molecule serves as a critical component during embryonic development from zygote formation through to blastocyst formation. During the early cell division stage siRNA ensures that gene expression and cellular differentiation remain controlled properly. The oocyte supplies maternal siRNAs that silence gene activity before embryonic transcription begins which enables the maternal-to-zygotic transition when the embryonic genome activates and maternal transcripts degrade.
- Maternal siRNA Contributions: Silencing Genes Before Embryonic Transcription
During the initial phase of embryogenesis maternal siRNAs function as crucial elements for gene silencing until embryonic transcription starts. The oocyte transfers siRNAs to the embryo which target maternal transcripts for degradation and facilitate the activation of the embryonic genome. Sperm siRNAs target and degrade the maternal transcript of SIRT1 which regulates MZT. The development and differentiation of the early embryo requires this particular process to proceed correctly.
siRNA and Body Patterning: Sculpting the Embryo
- Hox Gene Regulation: siRNA Fine-Tunes the Body Axis Blueprint
siRNA serves as a vital mechanism for controlling Hox genes that determine body axis formation throughout embryonic development. The Hox gene family comprises transcription factors that regulate the animal body axis from anterior to posterior. SiRNA enables precision control of gene expression required for correct morphogenesis and tissue differentiation. Research demonstrates that siRNA targeted at particular Hox genes can alter their expression levels to achieve accurate developmental process control.
Hox proteins Deformed (Dfd) and Ultrabithorax (Ubx) control different body structures throughout the anterior-posterior axis in Drosophila. While these proteins possess DNA binding capabilities that are alike they connect to separate genomic regions demonstrating the intricate nature of Hox gene control. By designing siRNA to target specific genomic regions researchers gain precise control over Hox gene activity which ensures correct patterning of the body axis.
- Gradient Formation: How Localized siRNA Creates Developmental Boundaries
The establishment of developmental boundaries during embryogenesis depends significantly on the creation of siRNA gradients. The ability of these gradients to establish sharp gene expression boundaries plays a critical role in proper morphogenesis. The distribution of small RNAs such as tasiR-ARFs establishes gradients that determine the adaxial-abaxial polarity in plant leaves. The accumulation gradient of tasiR-ARF across the leaf axis establishes a distinct boundary in ARF3 expression which demonstrates the role of siRNA gradients as positional markers that define developmental limits. Body patterning in vertebrates appears to involve similar mechanisms that use siRNA gradients. The development of somites in vertebrate embryos requires the integrated control of signaling pathways together with gene expression gradients. The precise spatial and temporal control of developmental processes needed for segmentation and body axis formation could be influenced by siRNA gradients.
- Comparative Insights
Research comparing Drosophila segmentation with vertebrate somite formation sheds light on siRNA's function in body patterning processes. The segmentation mechanism in Drosophila depends on the periodic activation of genes such as even-skipped and fushi tarazu through regulation by maternal and zygotic siRNAs. SiRNAs enable correct segmentation by turning off genes prior to the start of embryonic transcription and demonstrate the crucial maternal role in early embryo development. The "clock and wavefront" mechanism directs somite formation in vertebrates through oscillatory gene expression of cyclical and delta-like 1 which siRNA gradients may control to maintain accurate segmentation and somite boundary development. Research comparing both systems reveals that siRNA maintains a fundamental role in body patterning across various organisms even though their developmental processes differ.
Organogenesis Unlocked: siRNA's Tissue-Specific Roles
- Neural Tube Closure: siRNA Prevents Spina Bifida-Like Defects
The potential of siRNA for preventing neural tube defects including spina bifida has been demonstrated to be significant. The neural tube formation during embryogenesis is essential because any disruption in this process results in serious developmental defects. The use of siRNA allows researchers to silence essential genes for neural tube closure which maintains correct morphogenesis while preventing developmental defects. Scientific research shows that siRNA aimed at specific genes related to neural tube closure successfully prevents spina bifida-like defects in animal studies.
- Cardiac Development: Silencing Regulators of Heart Chamber Formation
siRNA functions as an essential component of cardiac development by targeting and silencing heart chamber formation regulators. Distinct cardiac chamber development requires meticulous genetic control mechanisms for proper formation. By designing siRNA molecules to target essential genes we can silence them which helps us understand the molecular mechanisms of cardiac development. The use of siRNA against fibronectin demonstrates its ability to interfere with epithelial branching during organ development which underscores its utility in cardiac morphogenesis modulation.
- Limb Bud Patterning: siRNA Gradients Digit Formation
The organization of digits within limb buds relies on essential siRNA gradients. Correct gene expression timing and placement during limb development stages determines successful limb formation. siRNA generates gradients which establish developmental boundaries necessary for proper digit formation. Research on Drosophila reveals that siRNA gradients serve as positional signals which establish developmental boundaries just as tasiR-ARFs function in determining plant leaf polarity. The preservation of this mechanism across different species indicates that siRNA gradients operate in the same manner during vertebrate limb development.
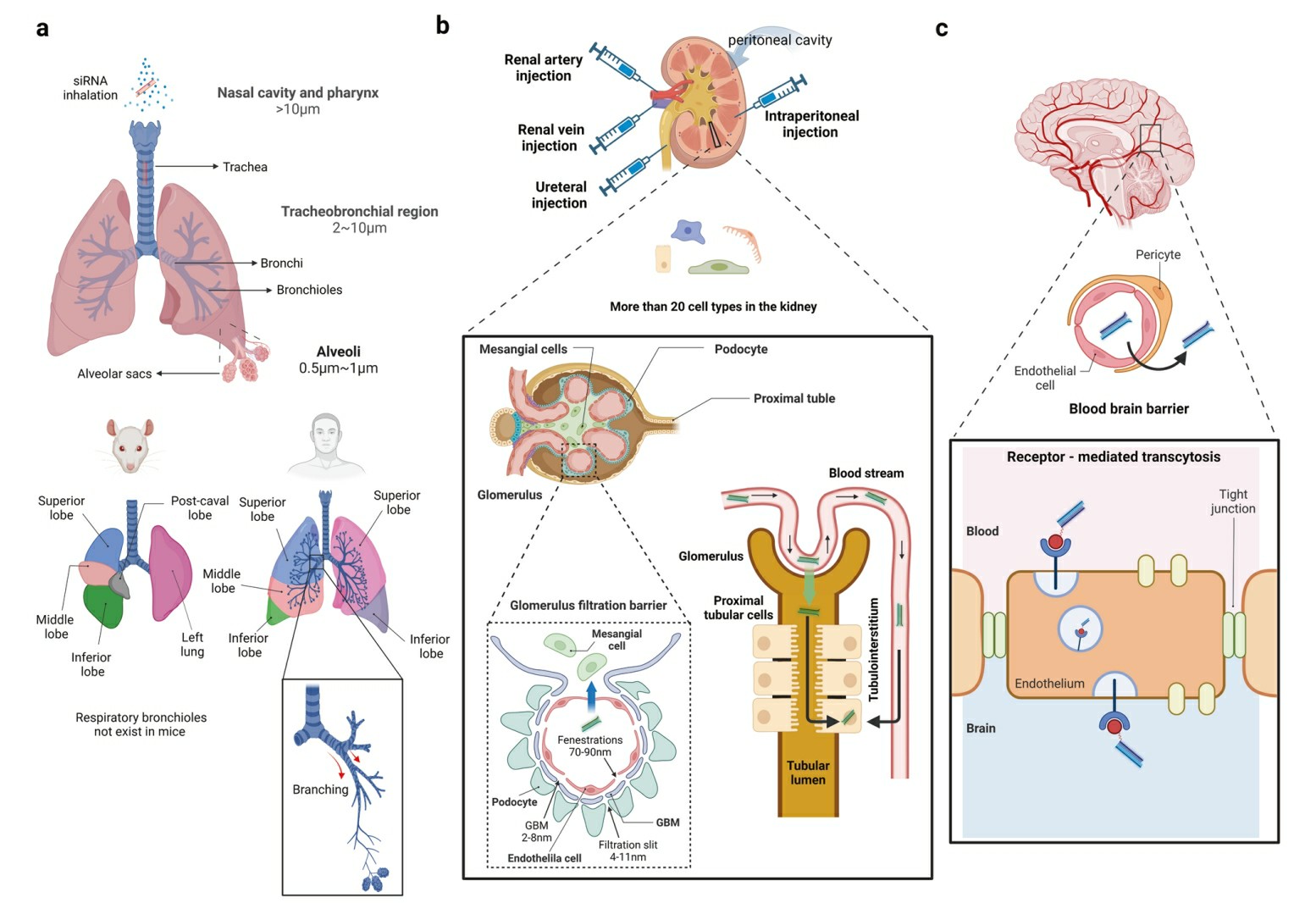 Fig. 1 Challenging organs to deliver siRNAs: lungs, kidneys, and brain1,6.
Fig. 1 Challenging organs to deliver siRNAs: lungs, kidneys, and brain1,6.
siRNA in Stem Cell Fate Decisions
- Pluripotency Maintenance: siRNA Knockdown of Differentiation Factors
Researchers have widely adopted siRNA to sustain stem cell pluripotency through the targeted silencing of differentiation-related genes. Research demonstrates that siRNA which targets specific differentiation-related genes can preserve the pluripotent state of embryonic stem cells (ESCs). A study discovered multiple essential genes including Tbx3, Esrrb, Tcl1, and Dapp4 which sustain pluripotency in mouse embryonic stem cells (mESCs). A genome-wide siRNA screen identified essential genes for mESC growth by showing that 68 genes when knocked down produced various phenotypes affecting both mESC viability and morphology. Research data demonstrates how siRNA technology can precisely adjust gene expression while preserving pluripotency.
siRNA Helps Choose Between Ectoderm/Mesoderm/Endoderm
siRNA determines stem cell fate by controlling transcription factors and signaling molecules that drive cells to differentiate into ectoderm, mesoderm, and endoderm. Research that employed RNAi to examine the transition from pluripotency to differentiation found multiple genes that control these biological functions. Research showed that siRNA targeting fibronectin interrupted epithelial branching during organ development which emphasizes its importance for lineage specification. Research findings show that siRNA can direct stem cells toward particular lineages by turning off genes that encourage different developmental paths.
- Organoid Applications: Using siRNA to Guide 3D Tissue Development
Organoids represent small-scale models of organs where siRNA demonstrates substantial potential for directing tissue development in three-dimensional structures. With siRNA treatment researchers can knock down genes that block particular differentiation courses enabling researchers to generate specific tissue types they desire. Research has shown that the delivery of SOX5, SOX6, and SOX9 genes through PEI-modified PLGA nanoparticles enhances chondrogenesis in human MSCs using siRNA technology. Scaffold-mediated siRNA delivery techniques have demonstrated the capacity to direct stem cell differentiation within implants which underscores the significance of siRNA for 3D tissue engineering applications. The demonstrated applications of siRNA in medical fields reveal its broad applicability for tissue engineering and regenerative medical solutions.
siRNA in Developmental Disorders: From Mechanism to Therapy
- Teratogen Protection: Using siRNA to Shield Against Birth Defects
siRNA displays therapeutic potential against teratogens responsible for birth defects by selectively silencing genes that drive the teratogenic process. Researchers used siRNA to fight against the development of microcephaly caused by the Zika virus (ZIKV). Scientists engineered small extracellular vesicles (sEVs) containing antiviral siRNA that targeted and inhibited ZIKV. The engineering process of these sEVs incorporated a neuron-specific RVG peptide derived from rabies virus glycoprotein to create neuro-specific targeting capabilities. The sEVsRVG-siRNA treatment in pregnant mice crossed both the placental and blood-brain barriers and reduced ZIKV infection in fetal brains alongside decreasing neuroinflammation and neurological damage.
- Congenital Disease Models: Correcting Mutations in Organoid Systems
Organoid models of congenital diseases allow scientists to correct genetic mutations using siRNA. A rat fetal model of spina bifida demonstrated therapeutic promise when researchers used intra-amniotic delivery of adenovirus-mediated siRNA against CRMP4 along with bone marrow stromal cell delivery. The treatment lowered CRMP4 expression while simultaneously encouraging neural differentiation and blocking cell apoptosis and repairing skin lesions in rat fetuses. The research showed how siRNA could boost BMSC transplantation outcomes for treating neural tube defects.
- Case Study: siRNA Therapy for Developmental Brain Disorders
In an in vitro study using human neuroblastoma cells (SHSY5Y), siRNA was used to silence the expression of three genes related to Alzheimer's disease (AD): Researchers used siRNA to target three Alzheimer's disease-related genes which included amyloid precursor protein (APP), tau protein, and voltage-dependent anion channel 1 (VDAC1). Decreased levels of target proteins led to enhanced synaptic activity and mitochondrial function which demonstrates that siRNA provides protection against AD. The HTT gene contains a trinucleotide expansion that causes Huntington's Disease (HD). Researchers used siRNA to target and silence the mutant HTT gene in laboratory and animal models. siRNA that targeted the upstream region of the CAG repeat in the HTT gene successfully suppressed its expression in cell cultures and showed potential for Huntington's disease treatment.
The earlier discussion highlighted that siRNA-loaded sEVs successfully inhibited ZIKV infection in mice showing microcephaly indicators. This strategy protected from viral infection transmission while reducing the neurological harm brought by ZIKV.
Comparative Developmental Biology: siRNA Across Species
- Evolutionary conservation: Core RNAi machinery from plants to mammals
The RNAi pathway together with essential siRNA machinery maintains high conservation throughout multiple species including plants and mammals. AGO proteins function as key components of the RISC while maintaining their presence across all eukaryotic organisms. In plants and animals AGO proteins attach to siRNAs or miRNAs which they then direct to complementary RNA targets to either degrade or repress translation of these targets. AGO proteins in plants cleave mRNA targets to control gene expression but in animals they mainly induce translational repression. The enzymes known as dicer process double-stranded RNA into siRNAs and exhibit conservation throughout different species. The production of siRNAs depends upon these enzymes which are necessary for RNAi pathway function. The fundamental process that drives siRNA-mediated gene silencing shows conservation. siRNAs bind to AGO proteins to form RISC that identifies and degrades complementary mRNA molecules. Both plants and animals utilize this mechanism as a defense strategy against viruses and transposons that attack cells.
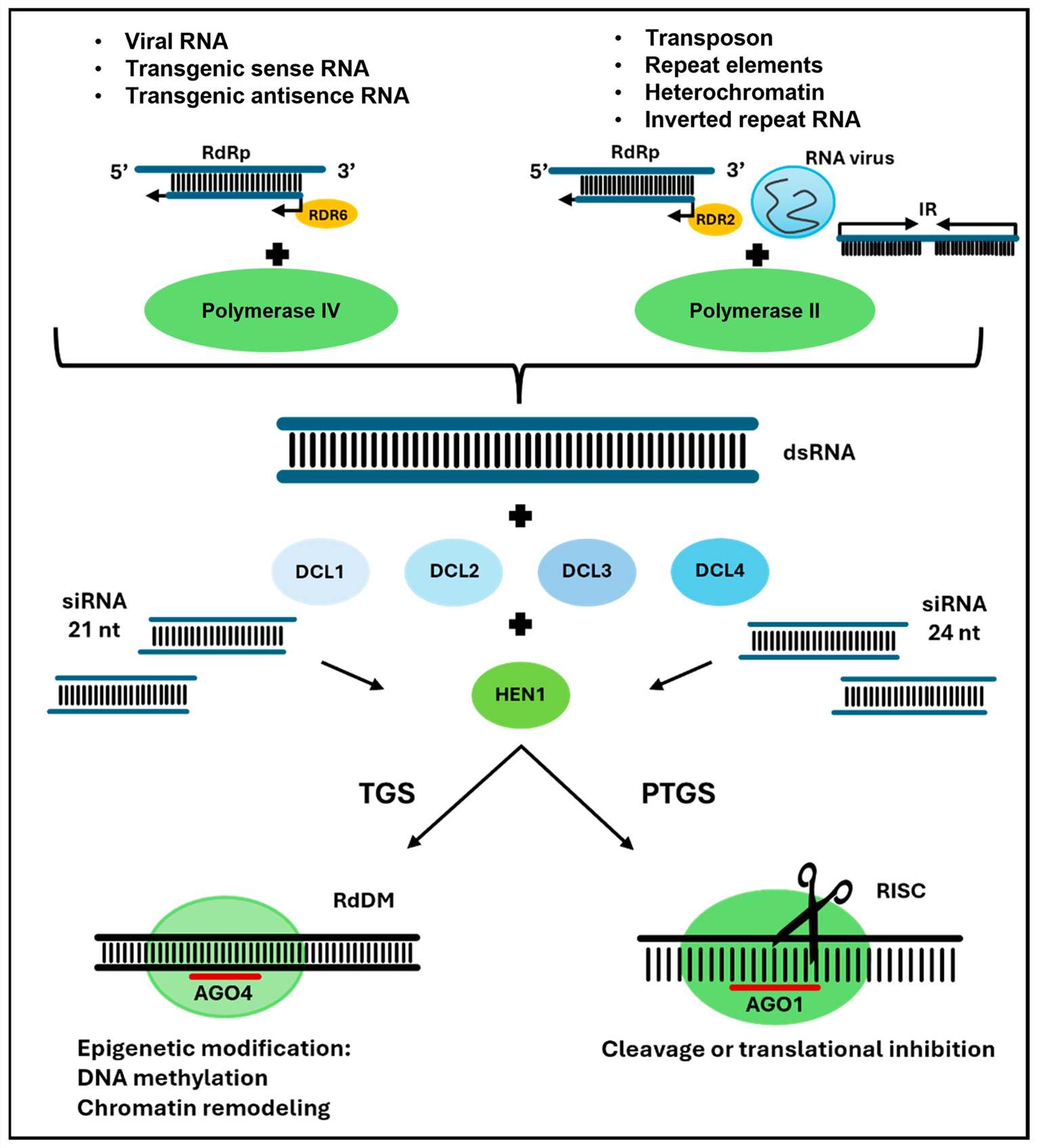 Fig.2 Mechanisms of the transcriptional gene silencing (TGS) or post-transcriptional gene silencing (PTGS) of siRNA-directed genes in plants2,6.
Fig.2 Mechanisms of the transcriptional gene silencing (TGS) or post-transcriptional gene silencing (PTGS) of siRNA-directed genes in plants2,6.
- Species-specific adaptations: Unique developmental roles for siRNA
siRNAs are essential for numerous developmental functions throughout plant systems. TAS3-derived tasiRNAs in Arabidopsis specifically target ARF3 and ARF4 genes which play critical roles in leaf polarity and development. A gradient of tasiRNAs is produced by localized expression of AGO7 and TAS3 which defines the adaxial (upper) and abaxial (lower) leaf sides. In shoot apical meristem development siRNAs function through the regulation of HD-ZIP III and WUSCHEL (WUS) genes to preserve stem cell populations.
Animals utilize siRNAs in various developmental functions. In mammals siRNAs function to control gene expression throughout embryonic development as well as tissue differentiation stages. Developmental gene expression regulation and genomic stability maintenance against transposable elements are functions of siRNAs in Drosophila.
Taxon-specific siRNAs have evolved in certain species to fulfill specialized functions within their development stages or adaptive processes. The Mimulus plant species evolved flower color by using taxon-restricted phased siRNAs that originated from an inverted duplication of a gene unrelated to the pigmentation pathway.
References:
- Ahn, Insook, Chanhee S. Kang, and Jinju Han. "Where should siRNAs go: applicable organs for siRNA drugs." Experimental & Molecular Medicine 55.7 (2023): 1283-1292. https://doi.org/10.1038/s12276-023-00998-y.
- Puchta-Jasińska, Marta, et al. "Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery." International Journal of Molecular Sciences 26.4 (2025): 1624. https://doi.org/10.3390/ijms26041624.
- Russell, Stewart J., et al. "An atlas of small non-coding RNAs in human preimplantation development." Nature Communications 15.1 (2024): 8634. https://doi.org/10.1038/s41467-024-52943-w.
- Pachamuthu, Kannan, Matthieu Simon, and Filipe Borges. "Targeted suppression of siRNA biogenesis in Arabidopsis pollen promotes triploid seed viability." Nature Communications 15.1 (2024): 4612. https://doi.org/10.1038/s41467-024-48950-6.
- Dubrovina, Alexandra S., and Konstantin V. Kiselev. "Exogenous RNAs for gene regulation and plant resistance." International journal of molecular sciences 20.9 (2019): 2282. https://doi.org/10.3390/ijms20092282.
- Distributed under Open Access license CC BY 4.0, without modification.
