Overview of DNAzymes
DNAzyme, also known as deoxyribozyme or catalytic DNA, is a class of enzymes whose chemical essence is short single-stranded DNA. Its special DNA sequence allows it to fold into a catalytically active structure in three-dimensional space and catalyze a series of biochemical reactions. Compared with enzymes composed of proteins or RNAs, DNAzymes benefit from their DNA chemical structure, have higher stability, and are easier to synthesize and amplify in vitro. Moreover, DNAzymes can still have the same or even better catalytic activity and specificity than other natural enzymes. Therefore, DNAzymes have important applications in biochemical fields such as biological imaging, detection, and sensing.
Classification and Functions of DNAzymes
DNAzymes have been divided into several classes according to their distinct functions. DNAzymes are normally isolated through in vitro selection methods from a random DNA library consisting of over 1000 DNA sequences. DNAzymes are DNA molecules that can perform catalytic functions. The catalytic activities of DNAzymes depend on their sequences and often require additional cofactors, such as metal ions and amino acids. DNAzymes can catalyze various chemical reactions, such as RNA cleavage, DNA cleavage by oxidation or hydrolysis, DNA/RNA ligation, and DNA phosphorylation. These features have been successfully applied in various biomedical applications, both in vitro and in vivo. By positioning the single nucleotide polymorphism at the recognition site, DNAzyme may perform allele-specific cleavage better than protein-based gene-silencing agents, which may be difficult for antisense oligonucleotide (ASO) or small interfering RNA (siRNA) due to thermodynamic restrictions on nucleotide hybridization and mismatch tolerance.
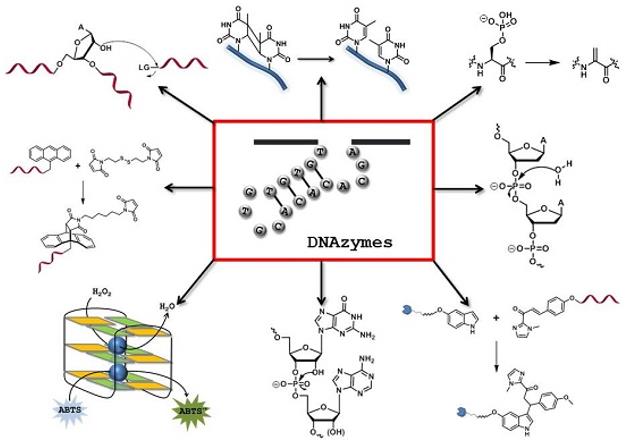 Figure 1. The Chemical Repertoire of DNAzymes. (Hollenstein M. 2015)
Figure 1. The Chemical Repertoire of DNAzymes. (Hollenstein M. 2015)
RNA-Cleaving DNAzymes
One of the most important properties of deoxyribozymes, which is also one of the most active research areas at present, is their function in cutting RNA molecules through esterification. This unique property of DNA makes it possible to destroy the RNA of cells and viruses in vivo, which has potential therapeutic effects in vivo. The most extensively investigated subgroup of DNAzymes are those that catalyze the cleavage of a single ribonucleotide (rA) linkage within a DNA strand. These DNAzymes are called RNA-cleaving DNAzymes. A representative RNA-cleaving DNAzyme is formed through the hybridization of a substrate strand and an enzyme strand, which consists of an active site, an enzymatic region, and two binding arms. The substrate strand has an rA linkage that acts as the cleavage site. When the cofactor is present, the enzyme strand forms a specific secondary structure that can cleave the rA linkage. This produces two fragments with a 2'-3' cyclic phosphate and a 5'-OH terminus, respectively. The DNAzyme's catalytic activity is highly specific to the substrate strand, and a single base mismatch in the antisense arms can significantly reduce the cleavage efficiency.
Preparation of DNAzymes
DNAzymes are not natural molecules, but they can be obtained from random-sequence DNA pools using in vitro selection. In vitro selection is a simple and powerful technique that can isolate rare DNA or RNA sequences with a desired function from a large population of single-stranded DNA or RNA molecules. DNAzymes are created by in vitro selection for catalytic functions. There are mainly two methods to obtain a functional DNAzyme. The first method involves the rational design of the DNAzymes' binding arms. However, the catalytic core of the DNAzyme must be known beforehand. The sequence of the binding arms can be designed similarly to primers for polymerase chain reaction (PCR), where their specificity will lead the DNAzyme to its target. The second method comes into place if the catalytic core is unknown. SELEX, or the Systematic Evolution of Ligands by Exponetial Enrichment, involves a largely random pool of nucleic acid. Any nucleic acid that can form a complex with the target molecule(s) is separated, amplified with PCR, and pooled together as a new library for another round of SELEX selection. Typically, 5–15 rounds of selection are performed, and the nucleic acids obtained at the end are the potential candidates. Next-generation sequencing (NGS) enables higher throughput sequencing of the final selection round products in fewer rounds than conventional Sanger sequencing. Instead of rationally designing DNAzymes that reuse known DNAzyme catalytic cores, SELEX identifies new types of DNAzymes and catalytic cores.
Application of DNAzymes
There is widespread interest in exploring DNAzymes for suitable applications, particularly in the area of biosensor research, which has been the subject of many previous reviews. There are many metal-ion-dependent RNA-cleaving DNAzymes, so it is not surprising that many biosensor studies are exploiting these DNA molecules as molecular-recognition elements. Back in 2000, the earliest example of using DNAzymes for biosensing was the design of a highly specific fluorescent biosensor for lead ions using a Pb2+-selective RNA-cleaving DNAzyme. Since then, using fluorescence as a platform to engineer RNA-cleaving DNAzyme biosensors has become a popular practice for two important reasons. First, fluorescence-based detection is highly sensitive and offers real-time reporting capabilities. Second, it is very convenient to turn an RNA-cleaving DNAzyme into a fluorescent sensor. In addition to the lead-ion sensor, fluorescent DNAzyme sensors for two other toxic metal ions, uranyl (UO22+) and mercury (Hg2+), have been successfully engineered. In addition, as a valuable tool in monotherapy or adjuvant therapy, DNAzymes continues to expand its therapeutic horizons in areas like gene editing (e.g., PNA-assisted DNAzymes to generate double-stranded breaks) and epigenetic modification (e.g., m6A vs. unmodified A at the cleavage site).
Reference
- Hollenstein M. DNA Catalysis: The Chemical Repertoire of DNAzymes. Molecules. 2015 Nov 20;20(11):20777-804. Distributed under Open Access license CC BY 4.0, without modification.
