siRNA in Metabolic Disorders-Silencing Genes to Restore Metabolic Balance
Introduction of siRNA in Metabolic Disorders
Small interfering RNA (siRNA) is a form of non-coding RNA involved in RNA interference (RNAi). SiRNA is approximately 21–23 nucleotides long, whcih binds to complementary mRNA and results in its degradation and inhibition of gene expression. It is used to silence genes associated with metabolic disorders, and RNAi inhibits expression of specific genes associated with metabolic disorders. This makes it more targeted than other methods, reducing the likelihood of off-target effects as it is complementary to only the specific gene. Binding of siRNA leads to the formation of the RNA-induced silencing complex (RISC), which causes degradation of mRNA, and ultimately silences the gene associated with it.
What are Metabolic Disorders
Metabolic disorders disrupt normal body metabolism including the processing and distribution of macronutrients like proteins, fats, and carbohydrates. Metabolic disorders can occur when abnormal chemical reactions in the body affect the normal metabolic process. It may also be defined as an inherited single gene disorder, most of which are autosomal recessive. Some symptoms that can occur with metabolic disorders are lethargy, weight loss, jaundice, seizures. The expressed symptoms would vary with the type of metabolic disorder. Metabolic disorders such as Obesity, Diabetes Type 2 (T2DM), and Inflammatory Bowel Diseases are the most prevalent worldwide. Inherited metabolic disorders are one cause of metabolic disorders, and occur when a defective gene causes an enzyme deficiency.
Design of siRNA for treatment of Metabolic Disorders
- GC Content
The role of the total GC content of an siRNA in determining its activity is still under debate. Although some papers suggest that an optimal GC content for an siRNA is 30–50%, other papers show that even highly active siRNAs with GC contents as high as 60% can be used. For this reason we advise the use of siRNAs with an overall GC content between 30 and 65% of base pairing nucleotides. It has been demonstrated that a GC stretch of 9 or more nucleotides anywhere in the base pairing sequence of an siRNA decreases its efficiency. Furthermore, siRNAs with a low GC content (less than two GC base pairs) in the 5' terminal third of the guide (antisense) strand are likely to be active siRNAs. These two features should be considered when designing siRNAs.
- Immune Stimulatory and Cytotoxic Sequence Motifs
Two of the siRNA sequence motifs capable of inducing strong immune response by binding to toll-like receptors are "GUCCUUCAA" and "UGUGU". In addition, "UGGC" is overrepresented in the guide strand of siRNAs with high cytotoxic activity. Therefore, the motifs should be avoided to reduce the cytotoxicity. siRNA target sites should be screened for similar sequences in the genome. There should be little or no sequence complementarity between the nontarget mRNAs and both strands of the siRNAs to prevent the use of siRNAs with off-target activity. To prevent off-target effects due to sequence redundancy, candidate siRNA sequences can be screened against EST databases, for example using BLAST. Importantly, any 19 nt random sequence can be screened against human EST databases and a significant number of hits with 15 or more matching nucleotides can be expected. However, not all mRNAs are expressed in the cell-line used in a given RNAi experiment. Hence, several siRNAs with fully independent sequences against a target gene should be used Controlling siRNA Off-Target Effects and should produce similar phenotypes.
- siRNA Strand Selection
SiRNAs are duplex RNAs and in principle both strands can be bound to Ago proteins as guide strands. However, only the guide (also called antisense strand) is functional against the target mRNA. The other strand (so-called passenger strand, often referred to as sense strand) is only active against non-target RNA. It is therefore imperative to create siRNAs with a maximum asymmetry for loading of the guide strand into the Ago protein. Two parameters are known so far, which allow the prediction of guide strand selection. Both endogenous and synthetic siRNAs are derived from double-stranded precursors and strand selection has to be determined. In general, strand selection follows the so-called asymmetry rule. The asymmetry rule states that the siRNA strand with the less stably paired 5' end will be selected and loaded into the Ago protein as the guide strand. The strand with the more stably paired 5' end will be mainly rejected as the passenger strand. When designing siRNAs the desired guide (antisense) strand should always have the less stably paired 5' end.
- Nucleotide Specificity
Structure studies of Ago proteins showed that the so called MID domain interacts with the 5' end of the guide strand. Due to its central location within the Ago protein the domain is named MID domain. Crystallization and KD studies of the MID domain with all four nucleotides in position 5 of the 5' end of the guide strand showed that uridine (U) binds with highest affinity to the MID domain, adenosine (A) with a decreased affinity and cytosine (C) and guanosine (G) with more than tenfold less affinity. Therefore, siRNA guide (antisense) strands should contain a U or A in position 5 of the 5' end. Avoid C and G. For the passenger (sense) strand 5' end, choose C and G to minimize strand incorporation. According to our unpublished data, the nucleotide specificity is not only a tool to control strand selection but siRNA strands with U or A at the 5' end also have a higher absolute affinity to Ago proteins and are therefore more potent siRNAs.
Avoiding siRNA Off-Target Activity by Experimental Design
- siRNA Concentration
The simplest, most economical and lowest cost method to reduce siRNA off-target activity is to use the lowest possible siRNA concentration. miRNA-like off-target effects and siRNA off-target effects caused by competition of siRNA- and miRNA-loading onto Ago proteins have been shown to be concentration-dependent. The first requirement for the use of low concentrations of siRNAs is the highest possible efficiency in siRNA delivery and activity. The design criteria described above may result in siRNAs with IC50 values in the range of 25–500 pM for unmodified siRNAs in cell-lines which are easily transfectable, e.g. HeLa, Cos-7, HEK293-T etc. In cell culture-based experiments using state-of-the-art transfection reagents, these siRNAs should achieve maximum on-target activity at concentrations <10 nM. To estimate IC50 values and the minimal concentration required for maximum siRNA activity we suggest to approximately titrate siRNA concentrations (use, e.g., 50; 10; 2; 0.4; 0.08 nM final siRNA concentration and compare to an unrelated siRNA and measure mRNA levels, e.g. by real-time PCR). We would not suggest using siRNAs for follow-up experiments if IC50 values >500 pM are determined in the case where cell-lines are used which can be transfected with high efficiency.
- Control siRNAs
In every RNAi experiment, some control siRNAs are needed. These can be siRNAs targeting a completely unrelated target-mRNA, which should not show any phenotype in the experimental context. Alternatively, random siRNA sequences or siRNAs targeting mRNA which are not expressed in the chosen biological system (e.g. siRNAs targeting Photinus luciferase or green fluorescent protein in cell-lines derived from human tissues) can be used. Note that as described under the topic "sequence redundancy", random siRNA sequences should have relatively high sequence complementarity (>15 nt) when compared to, e.g., human EST databases. Thus, two or more siRNAs should be used as unrelated control siRNAs. As mentioned before, the control siRNA-treated biological system should behave as closely as possible to untransfected or transfection reagent-treated biological system. As for the higher siRNA concentrations (>10–20 nM), control siRNAs will also induce off-target effects. For example, when control siRNAs are used at higher concentrations, often cell proliferation is decreased, compared to untransfected cells. Furthermore, control siRNAs should induce the same effects as very inefficient on-target siRNAs (e.g. <20–30% knockdown efficiency). In general, control siRNAs have to be designed according to the same principles as on-target siRNAs. This is particularly important with respect to siRNA/miRNA competition for Ago proteins.
siRNA Applications in Metabolic Disorders
- Hypercholesterolemia-siRNA Targeting PCSK9
Hypercholesterolemia is defined as LDL-C concentrations that are above the normal range. LDL-C increases the risk of cardiovascular diseases. Proprotein convertase subtilisin/kexin type 9 (PCSK9) plays a role in LDLR degradation, and thus affects the plasma levels of LDL-C by lowering the removal of LDL-C. Knockdown of PCSK9 by siRNA results in the reduction of PCSK9, which increases LDLR levels and reduces LDL-C. Inclisiran is a siRNA therapeutic agent that reduces LDL-C in humans. The level of LDL-C was reduced by 52% after 18 months treatment with inclisiran in the phase 3 clinical trial, and maintained a decrease at 24 months. Inclisiran is approved for the treatment of hypercholesterolemia and is considered safe. Future research can focus on the delivery system of siRNA for inclisiran, and on different target cell types and the combination of other lipid-lowering agents.
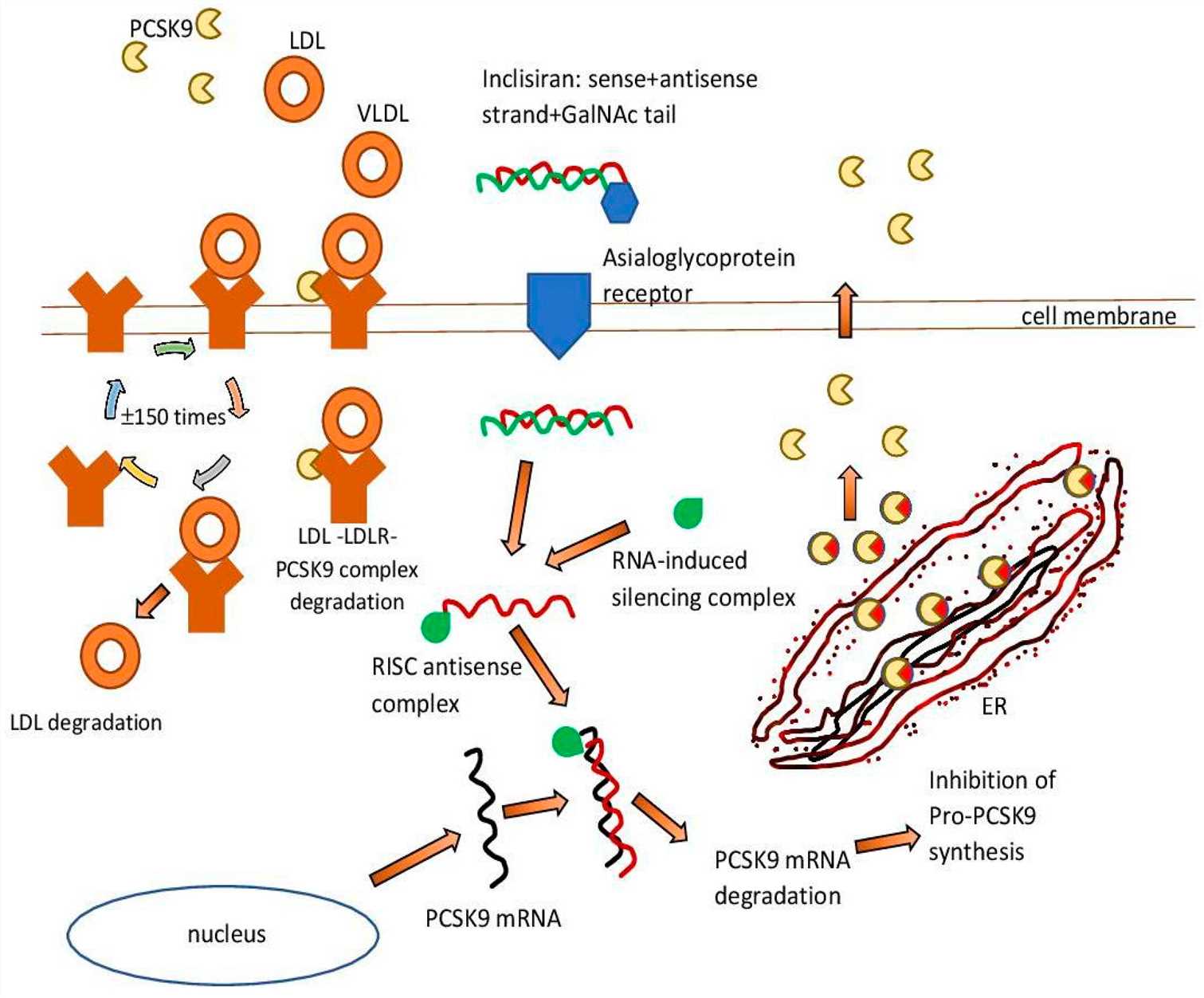 Fig. 1 Inclisiran—mechanism of action.1,6
Fig. 1 Inclisiran—mechanism of action.1,6
- Type 2 Diabetes-siRNA Targeting Glucagon Receptor (GCGR)
Type 2 diabetes is characterized by both insulin resistance and altered glucose homeostasis. The glucagon receptor (GCGR) controls the homeostasis of blood glucose levels via hepatic induction of gluconeogenesis and glycogenolysis. Reduced expression of GCGR via siRNA inhibits the action of the glucagon pathway, resulting in reduced hepatic production of glucose and increased insulin sensitivity. Reduction of GCGR expression via siRNA has been shown to lower blood glucose levels and improve glucose tolerance in diabetic animal models. For example, siRNA-mediated downregulation of GCGR expression reduced plasma glucose levels by 40% in diabetic mice and was effective for several weeks. GCGR siRNA is in preclinical development as a novel treatment for type 2 diabetes. Upcoming research will focus on enhancing siRNA sequence specificity and creating delivery systems that decrease off-target effects .
Current Status and Future Directions
- Current status
SiRNA has been explored for a range of metabolic disorders and several of them are now in advanced stages of clinical development. For example, ALN-KHK is an siRNA targeted to the ketohexokinase (KHK) gene and is a key component of fructose metabolism. High levels of fructose metabolism have been linked to metabolic syndrome and NAFLD, thus KHK is an attractive target for the development of siRNA therapeutics. Advanced clinical trials include several other siRNA therapeutic drugs besides ALN-KHK. The PCSK9-targeted siRNA medication Inclisiran caused substantial LDL-C reductions in hypercholesterolemia patients. It was also well tolerated in patients not controlled on maximal statin therapy, indicating it may serve as an adjunct to hypercholesterolemia. Another example is lumasiran which was approved by the FDA for the treatment of PH1. PH1 is a rare metabolic disorder where there is overproduction of oxalate and lumasiran targets the mRNA encoding glycolate oxidase (GO) which lowers oxalate production and helps alleviate the symptoms of PH1.
- Future Directions
Another critical issue for siRNA therapeutics is efficient and targeted delivery of siRNA to target tissue. Current delivery systems, such as LNPs, have shown some success but have limitations in terms of toxicity and specificity of tissue targeting. Future research will focus on improving the efficiency and safety of delivery vehicles for siRNA. Other delivery vehicles such as viral vectors (e.g., adeno-associated virus (AAV) and lentiviral vectors) have been shown to be promising for siRNA delivery. Viral vectors are able to transduce the target cells and lead to sustained expression of siRNA. Issues concerning immunogenicity and long-term safety have yet to be overcome. Research will focus on optimizing viral vectors to reduce the immune response and improve the therapeutic effects. In addition to the delivery vehicle, chemical modification of siRNA molecules can improve the stability of siRNA and reduce off-target effects. Examples of siRNA chemical modifications that can improve the stability and decrease off-target effects include 2'-O-methylation and phosphorothioate linkages. Research in this area will continue to explore novel chemical modifications that can improve the performance of siRNA.
References
- Wołowiec, Łukasz, et al. "Inclisiran—safety and effectiveness of small interfering RNA in inhibition of PCSK-9." Pharmaceutics 15.2 (2023): 323. https://doi.org/10.3390/pharmaceutics15020323.
- Gu, Wei, et al. "Polyphenols alleviate metabolic disorders: the role of ubiquitin-proteasome system." Frontiers in Nutrition 11 (2024): 1445080. https://doi.org/10.3389/fnut.2024.1445080.
- Kosmas, Constantine E., et al. "Inclisiran: a new promising agent in the management of hypercholesterolemia." Diseases 6.3 (2018): 63. https://doi.org/10.3390/diseases6030063.
- Kitamura, Hiroshi. "Ubiquitin-specific proteases (USPs) and metabolic disorders." International journal of molecular sciences 24.4 (2023): 3219. https://doi.org/10.3390/ijms24043219.
- Zhang, Yanzhen, et al. "Inclisiran: a new generation of lipid-lowering siRNA therapeutic." Frontiers in Pharmacology 14 (2023): 1260921. https://doi.org/10.3389/fphar.2023.1260921.
- Distributed under Open Access license CC BY 4.0, without modification.
