T-cell receptor (TCR) clonality assessment offers significant value in both clinical settings and research. Each T cell normally carries a unique TCR under healthy conditions. These receptors can recognize different antigens. This creates a highly diverse TCR repertoire in the body. Disease changes this pattern dramatically. One or a few T cell clones start to multiply rapidly during illness. This pathological expansion shows specific characteristics. The TCR repertoire becomes dominated by single clones or a few clones. Diversity drops significantly. These changes work as important molecular markers. Doctors use them for diagnosis, monitoring, and predicting outcomes. Clinically, TCR clonality tracking helps in multiple ways. It allows patient stratification based on immune profiles. It predicts how patients might respond to treatment. It monitors minimal residual disease (MRD) levels. It also measures how well immunotherapies are working.
At Creative Biolabs, we provide specific and reliable measurements of clonal diversity in T cell populations. Through this assessment, researchers can detect dynamic biomarkers across many diseases and understand the current state of the immune system.
TCR is a membrane protein made of two different chains. Most circulating T cells (90%-99%) carry receptors with α and β chains. The γ/δ types prefer epithelial tissues. The main job of αβ TCRs is recognizing foreign peptides alongside major histocompatibility complex (MHC) molecules.
The huge diversity of TCR repertoires forms the backbone of adaptive immunity. This diversity lets our immune system recognize almost unlimited numbers of antigens. The diversity comes from T-cell development in the thymus. During this process, genes rearrange through V(D)J recombination.
Gene segments combine during development. V (variable) and J (joining) segments create the variable part of the α chain. The β chain needs three segments: V, J, and D.
In mice, there are roughly 100 V and 60 J segments for α chains. For β chains, there are 35 V, 12 J, and 2 D segments. But about one-third of these don't work properly.
The most important part for antigen recognition is CDR3. This region recognizes the peptide-MHC complex. CDR3 sits at the V(D)J junction. Random nucleotides get added or removed between segments here. This creates even more diversity beyond what comes from basic recombination1.
Each new T cell gets a potentially unique receptor. This creates an extremely diverse TCR collection.
The thymus refines this diversity during T-cell maturation. Two selection processes occur here. Positive selection ensures MHC restriction. Negative selection creates central tolerance. Both processes are essential for proper T-cell function.
Scientists still debate the exact size of human αβ TCR repertoires. Estimates range from 25 million to 100 million unique receptors. In house mice, researchers found about 1.9 million unique TCR αβ combinations. The number of unique TCRβ chains reaches 500,000 to 800,000 different types1.
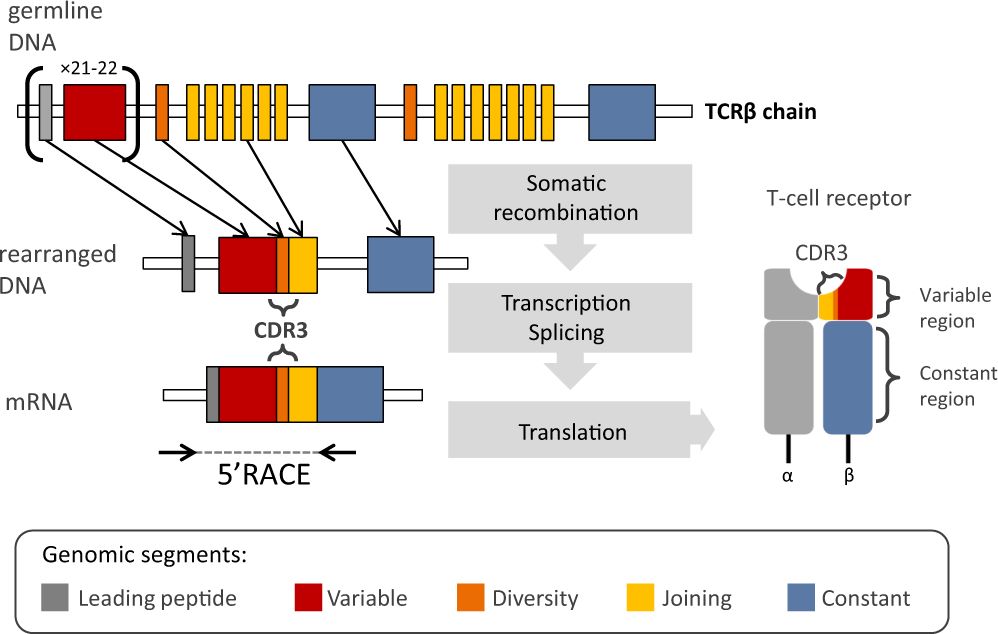 Figure 1 Schematic representation of the TCRβ chain rearrangement 2
Figure 1 Schematic representation of the TCRβ chain rearrangement 2
Disease changes this normal pattern. Chronic infections, cancer, or autoimmune conditions create persistent antigen exposure. This leads to sustained, large-scale expansions of specific clones. The balanced repertoire shifts toward oligoclonality, where just a few clones dominate.
T-cell cancers represent the extreme case. A single transformed cell grows uncontrollably, creating a monoclonal population that crowds out normal diversity.
These expanded clones fall into two categories:
Private clones are unique to each person. They reflect individual immune histories and genetic backgrounds.
Public clones appear across different people. These clones often share identical or very similar CDR3 sequences. They typically develop in response to common pathogens that many people encounter.
Understanding whether clones are private or public helps researchers interpret immune responses and disease patterns across populations.
Researchers have several ways to measure TCR clonality. The methods got much better over time. You pick one based on what you need to find out, how sensitive the test needs to be, what samples you have, your budget, and how fast you need answers.
| Method | Technology | Sample Requirements | Results/Output | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Spectratyping (CDR3 Length Analysis) | Capillary electrophoresis to separate fragments by size | RNA or cDNA samples |
Distribution plots:
|
|
|
| Multiplex PCR (BIOMED-2) | Amplifies TCR genes, then checks size and shape of products | Moderate amount of DNA | Shows distinct clonal peaks or polyclonal patterns |
|
|
| Flow Cytometry (TRBC1/VB) | Uses antibodies to identify T-cells with restricted TCR families | Fresh blood, bone marrow, or single-cell tissue preparations |
|
|
|
| NGS (Next-Generation Sequencing) | Sequences millions of TCR rearrangements simultaneously |
Very small amount of DNA/RNA Blood or archived tissue |
|
|
|
| Third-Generation Sequencing (Nanopore) | Reads long DNA pieces for complete V(D)J sequences | DNA or RNA samples | Full-length sequences |
|
|
What you choose depends on your situation:
Need quick answers? Flow cytometry works great
Want proven methods? BIOMED-2 PCR is reliable
Need detailed information? NGS tells you everything
Working outside the lab? Nanopore travels well
Tight budget? Spectratyping costs least (but barely works anymore)
| Workflow Stage | Service Component | Description |
|---|---|---|
| Library Preparation | DNA/RNA Extraction | Extract high-quality genetic material from different sample types |
| Template Preparation | Methods compatible with old tissue samples, fresh tissue, and liquid biopsies | |
| Quality Control | Comprehensive quality checks before proceeding to next step | |
| Target Enrichment | Multiplex PCR Amplification | Simultaneous amplification of all TCR gene segments (α, β, γ, δ) |
| Primer Design Optimization | Custom primer sets covering all V(D)J combinations | |
| Bias Reduction Technology | Minimized amplification bias for accurate quantification | |
| Sequencing Platform | NGS | High-throughput Illumina systems |
| TCR Sequencing | 10x Genomics and advanced platforms for individual cell analysis | |
| Real-time Sequencing | Oxford Nanopore for long DNA reads | |
| Customized Sequencing Depth | Adjustable coverage based on clinical case requirements |
| Development Area | Description |
|---|---|
| Algorithm Innovation | Proprietary computational programs for clonality data analysis |
| Assay Optimization | Continuous improvement of test sensitivity and accuracy |
| Custom Protocol Development | Tailored solutions for specific research needs |
| Technology Integration | Multi-platform combination for comprehensive analysis |
| Application Category | Specific Application | Purpose |
|---|---|---|
| Hematologic Malignancy Diagnosis | T-cell Lymphoma/Leukemia | Diagnosis and classification |
| Minimal Residual Disease (MRD) | Disease monitoring | |
| Relapse Detection | Early warning system | |
| Immunotherapy Monitoring | CAR-T Cell Therapy | Treatment efficacy assessment |
| Immune Checkpoint Inhibitors | Response prediction | |
| Post-transplant GVHD | Risk evaluation | |
| Autoimmune Disease Research | Disease Activity Assessment | Monitor disease progression |
| Treatment Response | Evaluate therapeutic effectiveness |
Researchers now rely on TCR clonality measurements for both patient care and research. This technique has found uses in many medical areas.
Blood cancers, particularly T-cell lymphomas, show a clear pattern. A single dominant TCR clone appears when the disease develops. This finding helps doctors confirm their diagnosis. NGS changed everything. Traditional PCR methods could only detect 44-72% of cutaneous T-cell lymphoma cases. NGS now catches 69-100% of them. Doctors combine TCR data with genetic mutations. This combination helps them predict how patients will respond to treatment. The approach works best in early mycosis fungoides cases.
NGS-based TCR sequencing detects cancer cells at extremely low levels. It can find one cancer cell among 100,000 healthy ones. Flow cytometry can only detect one in 1,000 to 10,000 cells. This sensitivity matters in AML. Doctors spot returning cancer much earlier. Early detection creates more treatment options.
TCR analysis shows whether immunotherapy is working. Successful treatments cause specific T-cell clones to multiply rapidly. This expansion often happens before patients show clinical improvement. HPV16-driven cancers provide a clear example. Patients who developed dominant TCR clones after chemoradiation achieved complete remission. This discovery led to "TCR-focused" therapies. Doctors now identify each patient's most effective anti-tumor TCRs. They use these TCRs to create personalized cell treatments.
TCR sequencing identifies the T-cell clones that attack healthy tissue. For example, in Lupus Nephritis, scientists found overactive PD-1⁺ CD8⁺ T-cell clones in inflamed kidneys. PD-1 pathway drugs might help these patients. In Primary Sjögren's Syndrome, researchers discovered GZMK⁺ CXCR6⁺ CD8⁺ T-cells in both blood and salivary glands. More of these cells means worse disease. IL-15 pathway treatments show promise. Among Autoimmune Hemolytic Anemia, patients with clonal T-cell populations get sicker. They also take longer to respond to treatment.
Some bone marrow transplant patients develop dangerously low blood counts. Large-granular lymphocytosis causes this problem. TCR analysis proves that clonal T-cell expansion drives the condition in nearly half of cases. These finding changes treatment decisions. Doctors use immunosuppressive drugs to solve the problem. The treatment works well.
TCR analysis reveals how our immune system fights infections and responds to vaccines. In Chronic Viral Infections: Viruses like Cytomegalovirus permanently alter our TCR patterns. They create lasting, oligoclonal signatures that researchers can track for over ten years. Among COVID-19 Research, scientists compared mRNA vaccination to natural infection using TCR sequencing. The immune responses differ in timing and strength. T-cell clones expand and shrink at different rates. This information helps design better booster strategies. The technique also tracks SARS-CoV-2-specific memory T-cells over time. Vaccination creates immune protection that lasts.
TCR clonality assessment moved from research labs to hospital bedrooms. It now helps diagnose diseases and predict outcomes. Doctors use it to monitor treatments and design personalized therapies. The technique works across many medical specialties.
Technical Excellence
We deliver results faster than most competitors in the market. Our testing maintains high accuracy standards with built-in quality guarantees. You don't need to worry about sample handling or data processing. We manage the entire workflow from sample receipt to final report delivery. This complete service approach saves you time and reduces potential errors.
Beyond Basic Testing
We offer custom bioinformatics analysis tailored to your specific research questions. Our team provides clinical trial support to help you meet regulatory requirements. We host free technical training sessions at academic conferences. You can also get publication support when you need help analyzing data for research papers. These extra services make us more than just a testing lab.
Our team handles the complex technical work so you can focus on patient care and research. We deliver reliable results quickly with the support you need.
When is TCR clonal testing needed?
This test is mainly used for patients suspected of having T-cell lymphoma. It's particularly useful when patients have enlarged lymph nodes or abnormal lymphocytes in blood. The test helps distinguish between benign and malignant conditions. It's also used to monitor treatment response and screen for disease recurrence.
How do I interpret the test results?
A positive result means T-cell clones were detected. This suggests possible malignant disease. A negative result usually indicates reactive hyperplasia. However, final diagnosis requires combining clinical symptoms, imaging studies, and other pathological tests.
What makes your testing service special?
We use high-throughput sequencing technology. Our test has high sensitivity and can detect low-frequency clones. We cover major TCR gene loci for accurate results. Our molecular diagnostic team provides detailed result interpretation and clinical recommendations.
References
For any technical issues or product/service related questions, please leave your information below. Our team will contact you soon.
All products and services are For Research Use Only and CANNOT be used in the treatment or diagnosis of disease.
 NEWSLETTER
NEWSLETTER
The latest newsletter to introduce the latest breaking information, our site updates, field and other scientific news, important events, and insights from industry leaders
LEARN MORE NEWSLETTER NEW SOLUTION
NEW SOLUTION
CellRapeutics™ In Vivo Cell Engineering: One-stop in vivo T/B/NK cell and macrophage engineering services covering vectors construction to function verification.
LEARN MORE SOLUTION NOVEL TECHNOLOGY
NOVEL TECHNOLOGY
Silence™ CAR-T Cell: A novel platform to enhance CAR-T cell immunotherapy by combining RNAi technology to suppress genes that may impede CAR functionality.
LEARN MORE NOVEL TECHNOLOGY NEW SOLUTION
NEW SOLUTION
Canine CAR-T Therapy Development: From early target discovery, CAR design and construction, cell culture, and transfection, to in vitro and in vivo function validation.
LEARN MORE SOLUTION