Deciphering the Molecular Mechanisms of Respiratory Syncytial Virus Infection
Respiratory Syncytial Virus (RSV) is a globally prominent respiratory pathogen, recognized as a leading cause of severe lower respiratory tract infections (LRTIs) in infants, young children, the elderly, and immunocompromised individuals. Its widespread prevalence and the substantial burden it places on global healthcare systems underscore the critical need for a deeper molecular understanding of its infection and pathogenesis. Research into these fundamental mechanisms is paramount, as it forms the necessary foundation for developing effective vaccines and targeted antiviral therapeutics.
RSV: The Viral Blueprint
RSV belongs to the Orthopneumovirus genus and is a non-segmented, negative-sense, single-stranded RNA virus. Its genome is approximately 15.2 kilobases (kb) in length and encodes 10 genes that express 11 distinct proteins.
The Core Components and Their Roles
The RSV virion possesses a lipid envelope and a complex internal structure. Key structural and non-structural proteins drive the infection process:
| Gene | Encoded Protein | Primary Function |
|---|---|---|
| NS1/NS2 | Non-structural proteins | Inhibits interferon signaling, facilitates immune evasion |
| N | Nucleocapsid protein | Encapsulates the genomic RNA, forming the ribonucleoprotein (RNP) complex |
| P | Phosphoprotein | Scaffold for the replication complex |
| M | Matrix protein | Essential for viral assembly and budding |
| G | Attachment glycoprotein | Mediates receptor binding and modulates the immune response |
| F | Fusion protein | Triggers membrane fusion, enabling viral entry and cell-to-cell fusion (syncytia formation) |
| L | Large polymerase | Possesses RNA-dependent RNA polymerase (RdRP) activity |
| M2-1/M2-2 | Transcription regulators | M2-1 promotes mRNA synthesis; M2-2 regulates replication/transcription switch |
| SH | Small hydrophobic protein | Modulates ion channels and can influence apoptosis |
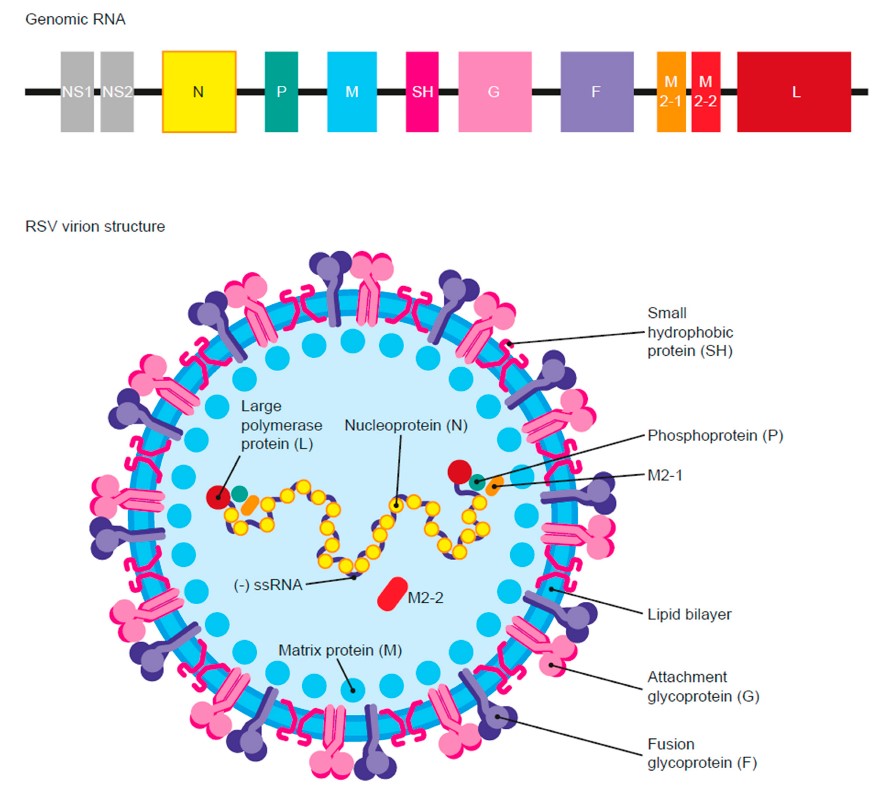 Fig.1 Respiratory Syncytial Virus (RSV) Genome and Virion Structure.1
Fig.1 Respiratory Syncytial Virus (RSV) Genome and Virion Structure.1
Services you may interested in
The Molecular Life Cycle of RSV
The RSV replication cycle is a precisely orchestrated sequence of molecular events occurring within the host cell cytoplasm.
Entry, Transcription, and Replication
- Attachment and Entry: The life cycle begins with the viral attachment protein, G protein, binding to an unknown host cell receptor. Following attachment, the F protein undergoes a critical conformational change, triggering a pH-independent fusion of the viral and cellular membranes. This fusion allows the viral core to enter the host cell cytoplasm.
- Transcription: Once inside, the viral RdRP (the L protein) starts transcribing the negative-sense genomic RNA into individual messenger RNAs (mRNAs). The M2-1 protein is essential as a processivity factor for this transcription.
- Replication: At a later stage, the RdRP switches to replication mode, producing a full-length, positive-sense intermediate known as the antigenome. This antigenome then serves as the template for synthesizing new, negative-sense progeny genomic RNA molecules.
Assembly and Release
The newly synthesized genomic RNA is encapsulated by the N protein to form new RNP complexes. The M protein is crucial in driving the assembly of new viral particles at the host cell membrane. The SH protein also plays a role in regulating the final budding process. Complete virions are then released through exocytosis to spread the infection to neighboring cells. Crucially, the F protein also mediates the fusion of infected cells with nearby uninfected cells, creating large, multi-nucleated cells called syncytia, which is a hallmark of RSV pathology.
Mechanisms of Pathogenesis: How RSV Causes Disease
RSV's ability to cause disease stems from both its direct cytopathic effects and its profound manipulation of the host immune system.
Immune Evasion and Antagonism
The non-structural proteins, NS1 and NS2, are key virulence factors and powerful immune antagonists. Their primary function is to suppress the host's innate antiviral response, particularly the interferon (IFN) signaling pathway. Specifically, NS1/NS2 are known to inhibit the production of type I interferons (IFN-β) and block signaling pathways like the RIG-I/MAVS and STAT1/2 pathways, effectively rendering the host cell defenseless against the virus.
Cellular Perturbation
RSV infection profoundly alters the molecular landscape of the host cell:
- Cellular Fusion: The F protein directly mediates cell-to-cell fusion, leading to the formation of syncytia, which allows the virus to spread efficiently while evading neutralizing antibodies.
- Cell Signaling Disruption: The virus activates cellular signaling molecules like NFkB and RhoA, leading to the release of inflammatory cytokines and chemokines.
- Cytoskeletal Remodeling: The virus induces the rearrangement of the actin cytoskeleton. This cellular manipulation facilitates both the efficient budding of new viral particles and the formation of the pathological syncytia.
The Dual Role of Surface Glycoproteins
The surface glycoproteins are major determinants of pathogenesis and critical targets for intervention.
- F Protein: Highly conserved across RSV strains, the Fusion protein (F) is the main target for neutralizing antibodies and a prime candidate for vaccine development due to its essential role in membrane fusion.
- G Protein: The Attachment glycoprotein (G) facilitates initial cell binding. Interestingly, it also functions as a chemokine mimic (specifically, CX3C chemokine mimicry), actively modulating the host immune cell chemotaxis and influencing the inflammatory profile of the infection. Along with the F protein, the G protein is also a preferred target for vaccine strategies.
Molecular Epidemiology and Diagnostics
A molecular understanding of RSV extends into its clinical and epidemiological aspects.
Strain Variability and Evolution
RSV is categorized into two major antigenic groups, A and B, which circulate globally, often concurrently. Within these groups, various genotypes exist (e.g., A group: ON1, GA2; B group: BA9, BA10). Tracking mutations, particularly in key proteins like the F protein (which can relate to antiviral resistance) and the G protein (changes in glycosylation sites), is essential for understanding seasonal trends and pathogenesis.
Molecular Detection
The gold standard for clinical diagnosis relies on highly sensitive molecular techniques:
- Real-time Reverse Transcription PCR (RT-qPCR): This technique specifically targets RSV RNA, providing high sensitivity and is the most common diagnostic method.
- Genotyping: Whole-genome sequencing or specific G protein gene PCR assays are used to quickly differentiate between A and B subtypes and track genotypic variations in the circulating strains.
Frontier Research in Molecular Virology
Ongoing research continues to unravel the finer details of RSV's molecular biology, utilizing sophisticated platforms to accelerate the development of countermeasures.
- Reverse Genetics Systems: These systems allow scientists to build recombinant RSV viruses with specific gene modifications, enabling precise functional studies of individual viral proteins and their contribution to pathogenesis.
- High-Resolution Structural Biology: Techniques like Cryo-Electron Microscopy (CryoEM) are providing unprecedented, high-resolution insights into the structure of critical complexes, such as the RdRP and its promoter, revealing potential new targets for antiviral drugs.
- Organoid Models: Human airway organoids are being used to simulate the virus-host interactions in vitro, offering a more physiologically relevant model than traditional cell culture for studying the mechanisms of infection.
In conclusion, the RSV virus is a highly evolved pathogen with a sophisticated molecular arsenal dedicated to efficient replication and potent immune evasion. The proteins F, G, NS1, and NS2 are central players in this molecular battle, serving as the main architects of viral entry, immune suppression, and disease pathology. Continued, rigorous exploration of these molecular mechanisms remains the most direct path to developing the next generation of effective vaccines and antiviral therapies to combat this significant global health threat.
If you want to learn more about the norovirus vaccine, please refer to:
Understanding the Molecular Mechanisms of Three RSV Vaccine Construction Strategies
Respiratory Syncytial Virus F Protein in Next-Generation Vaccine Design
Browse our Norovirus Antigen Products
Need a custom solution? If our off-the-shelf products aren't a perfect fit, we can create one for you. Contact us to design a product that precisely matches your experimental demands.
References
- Jenkins, Victoria A., et al. "The quest for a respiratory syncytial virus vaccine for older adults: thinking beyond the F protein." Vaccines 11.2 (2023): 382. https://doi.org/10.3390/vaccines11020382
- Distributed under Open Access license CC BY 4.0, without modification.
Created October 2025
Frequently Asked Questions (FAQs)
Q: What is the length of the RSV genome?
A: The RSV genome is approximately 15.2 kb in length and consists of negative-sense, single-stranded RNA.
Q: Which RSV proteins are the primary targets for vaccine development?
A: The F protein and G protein are the main targets for vaccine development. The F protein is highly conserved and induces potent neutralizing antibodies, while the G protein mediates viral attachment to host cells and is involved in immune modulation.
Q: How can RSV subtype A and subtype B be distinguished?
A: RSV subtypes A and B can be differentiated using molecular methods, such as PCR with subtype-specific primers targeting the G protein gene or full-genome sequencing. The G protein gene exhibits significant genetic differences between the two subtypes, making it an ideal target for differentiation.
Q: What is the main impact of NS1 and NS2 proteins on the host immune system?
A: The NS1 and NS2 proteins inhibit the host's antiviral immune response by blocking the production of type I interferons (IFN-α/β) and disrupting STAT1/2 signaling. This prevents the activation of IFN-stimulated genes, allowing RSV to replicate unchecked.
Q: What is the most commonly used molecular diagnostic method for RSV in clinical practice?
A: Real-time RT-PCR (quantitative RT-PCR) is the gold standard for clinical RSV detection. It is highly sensitive, specific, and fast, allowing for the rapid identification of RSV in patient samples.
All of our products can only be used for research purposes. These vaccine ingredients CANNOT be used directly on humans or animals.


