Clier-qPCR:Quantitative Validation of GlycoRNAs
GlycoRNAs, newly identified RNA species modified by glycans, represent an exciting frontier bridging glycobiology and RNA biology. Their detection and quantification demand techniques that combine sensitivity, specificity, and quantitative power. Clier-qPCR (click chemistry-based enrichment of glycoRNAs RT-qPCR) emerges precisely at this intersection. Leveraging click chemistry bioorthogonal reactions, Clier-qPCR enables specific enrichment and real-time quantitative analysis of glycoRNAs in various biological contexts. At Creative Biolabs, we offer specialized services tailored to these needs. Our Clier-qPCR for quantitative validation of glycoRNAs service combines the specificity of click chemistry with the sensitivity of real-time PCR, enabling accurate validation of glycoRNA candidates. Complementing this, our comprehensive glycorna analysis technologies suite provides advanced tools for glycoRNA profiling, glycoRNA functional analysis, and imaging for glycosylated RNA. With our expertise, researchers can confidently explore the roles of glycoRNAs in various biological processes.
What is Clier-qPCR?
Clier-qPCR integrates click chemistry-based enrichment with real-time quantitative PCR (RT-qPCR) to specifically validate and quantify glycoRNAs. It relies on the biotinylation of glycoRNAs via click chemistry reactions such as DBCO-biotin conjugation, followed by streptavidin magnetic bead capture and downstream qPCR analysis.
| Feature | Description |
|---|---|
| Reaction type | Click chemistry (DBCO-azide cycloaddition) |
| Enrichment method | Streptavidin bead capture of biotin-labeled RNAs |
| Quantification method | RT-qPCR (ΔΔCt analysis) |
| Target molecules | Glycosylated RNAs (glycoRNAs) |
| Applicable RNA size range | 50–2000 nucleotides |
Why Use Clier-qPCR?
Clier-qPCR specifically validates candidate glycoRNAs discovered through methods like Clier-seq. Its applications include:
- Verification of glycoRNA candidates.
- Quantitative expression analysis across cell types, tissues, and conditions.
- Functional studies linking glycoRNA presence to biological processes.
Key Advantages of Clier-qPCR
| Advantage | Details |
|---|---|
| High specificity | Click chemistry ensures selective labeling of glycoRNAs. |
| High sensitivity | Detects low-abundance glycoRNAs. |
| Broad coverage | 50–2000 nt RNAs, encompassing lncRNAs, sncRNAs, pre-tRNAs. |
| Dual function | Validates presence and quantifies expression. |
| Reduced false positives | Negative controls distinguish specific from nonspecific enrichment. |
| Efficiency | One-day workflow from RNA to data. |
Detailed Workflow of Clier-qPCR
Step 1: Sample Preparation
- (Optional) Incubated with Ac4ManNAz for 48-72h
- Extract total RNA with trizol, purify, and treat with DNase I and proteinase K.
Step 2: Click Chemistry Labeling
- Incubate RNA samples with a biotinylated cyclooctyne derivative under controlled temperature conditions to facilitate selective labeling.
Step 3: GlycoRNA Enrichment
- Capture biotin-labeled RNAs with streptavidin magnetic beads.
- Retain a portion as input control.
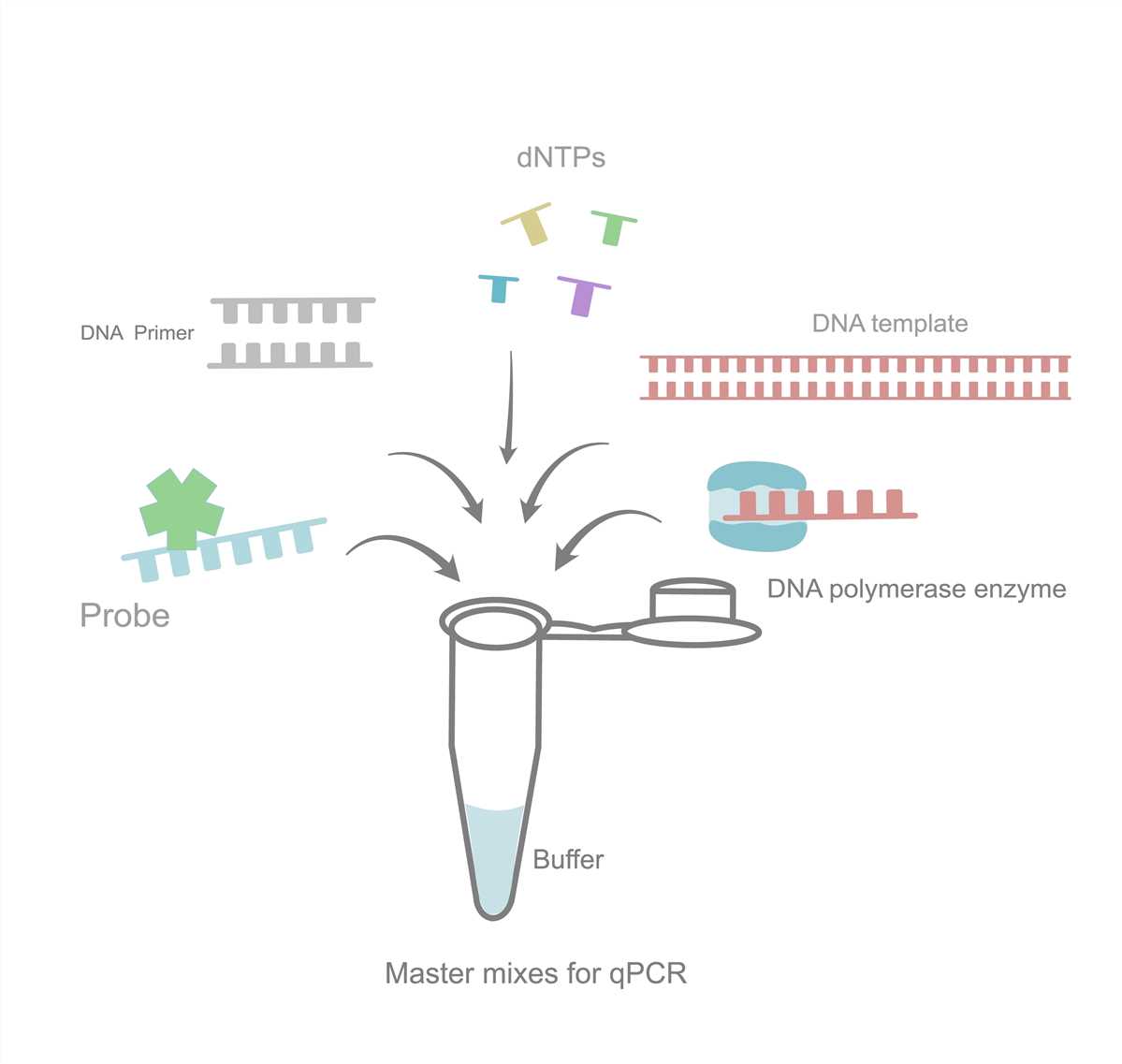
Step 4: cDNA Synthesis
- Use cDNA synthesis kit to synthesize cDNA.
- Random primers and dNTPs included.
Step 5: RT-qPCR Detection
- Use commercialized fast reagent kits and target-specific primers.
- Calculate enrichment fold by ΔΔCt method.
Experimental Design Considerations
| Category | Details |
|---|---|
| Sample Selection | Various cell lines, tissues, treated/untreated groups |
| Control Groups | Unlabeled and input controls |
| Primer Design | Target-specific primers and internal reference genes |
| Data Analysis | ΔΔCt calculation, t-tests or ANOVA for significance |
Sample Requirements for Clier-qPCR
| Sample Type | Requirement | Other Quality Requirements |
|---|---|---|
| Cells | 1×10⁶–10⁷ cells |
|
| Tissue | 10–50 mg fresh or frozen | |
| Body Fluids | 1–5 mL (blood, saliva, urine) |
Applications
Disease Research
- Cancer: GlycoRNAs as diagnostic markers (e.g., tumor-associated lncRNAs).
- Neurodegeneration: Changes in glycoRNA profiles in Alzheimer's models.
- Immunology: Role of glycoRNAs in immune activation and inflammation.
Drug Development
- Target Identification: GlycoRNAs as novel druggable targets.
- Therapeutic Response Monitoring: Changes in glycoRNA profiles post-treatment.
Basic Research
- RNA Biology: Mechanisms and dynamics of RNA glycosylation.
- Gene Regulation: GlycoRNA impact on transcriptomic regulation.
Published Data
Case 1: Validation of Novel GlycoRNAs
High-throughput Clier-seq analysis identified several unannotated long non-coding RNAs (lncRNAs) in P3HR1, Akata, and CNE2 cell lines, including MSTRG.7832, MSTRG.5930, and MSTRG.18836. To confirm their glycosylation status, Clier-qPCR was employed, comparing cells labeled with Ac4ManNAz (Ac4+) to unlabeled controls (Ac4−). The results demonstrated significant enrichment (>200-fold) of these RNAs in the Ac4+ groups, with minimal signals in the Ac4− controls. For instance, MSTRG.7832 in P3HR1 cells exhibited a 250±15-fold enrichment. These findings validated the glycosylation of these novel lncRNAs, subsequently named LINC03118, LINC03119, and USP43-OT.
Case 2: Quantitative Analysis of GlycoRNA Across Cell Lines
The glycosylation levels of Vault RNA vtRNA2-1 were assessed across P3HR1, CNE2, and Akata cell lines using Clier-qPCR. P3HR1 cells showed the highest glycoRNA enrichment (120±8-fold), followed by CNE2 cells (90±6-fold), while Akata cells exhibited minimal enrichment (<20-fold). These differences correlated with the relative expression levels of vtRNA2-1 mRNA, suggesting that glycosylation is influenced by both expression levels and cell-type-specific factors.
Case 3: Glycosylation Patterns in Precursor tRNAs
Clier-qPCR was utilized to investigate the glycosylation status of precursor and mature tRNAs. The analysis revealed that precursor tRNAs exhibited higher glycoRNA enrichment compared to their mature counterparts. For example, pre-tRNA-Lys-TTT-7-1 showed a 300±20-fold enrichment, whereas the mature tRNA displayed a 150±10-fold enrichment. These results suggest that glycosylation occurs during tRNA maturation, highlighting a potential regulatory mechanism in RNA processing.
Accurate measurement and detection remain crucial fundamental requirements in the fast-paced field of glycoRNA research. The Clier-qPCR system provides researchers with precise, sensitive, and versatile tools for validating and quantifying glycoRNA. The integrated platform utilizes click chemistry reactions together with real-time PCR sensitivity to generate novel investigative methods for RNA glycosylation biology and its applications. Creative Biolabs provides essential support for both novel glycoRNA candidate validation and comprehensive analytical research. Please reach out to us to learn more about our services or to discuss how we can adapt our services to match your research requirements.
Reference:
- Zhu, Nannan, et al. "Transcriptome-wide Identification of GlycoRNAs by Clier-seq Pipeline." bioRxiv (2025): 2025-02. https://doi.org/10.1101/2025.02.28.639800