GlycoRNA Imaging Service
Introduction to GlycoRNA
GlycoRNAs are RNA molecules modified by glycans (like sialic acid or GlcNAc) found on cell surfaces. These molecules play key roles in immune regulation, cell signaling, and disease development. The glycoRNA synthesis process is dynamic within a cell. It begins in the endoplasmic reticulum, where a precursor oligosaccharide interacts with γ–RNA to form glycoRNA. After that, the nascent glycoRNA then moves via vesicles to the Golgi apparatus. In the Golgi, further modifications occur, which are integral parts of the dynamic synthesis process. These modifications refine the glycoRNA structure. Finally, the modified glycoRNA is transported in vesicles to the plasma membrane. Throughout this journey, from the endoplasmic reticulum to the Golgi and then to the plasma membrane, the synthesis of glycoRNA is a continuously evolving and dynamic process, and their low abundance and dynamic nature make them challenging to study with traditional methods. Creative Biolabs offers glycoRNA imaging service, a cutting-edge solution to visualize and analyze glycoRNA interactions in real time. Using advanced glycoRNA profiling solutions, imaging technologies, and functional analysis for glycosylated RNA, we help researchers uncover the roles of glycoRNAs in health and disease.
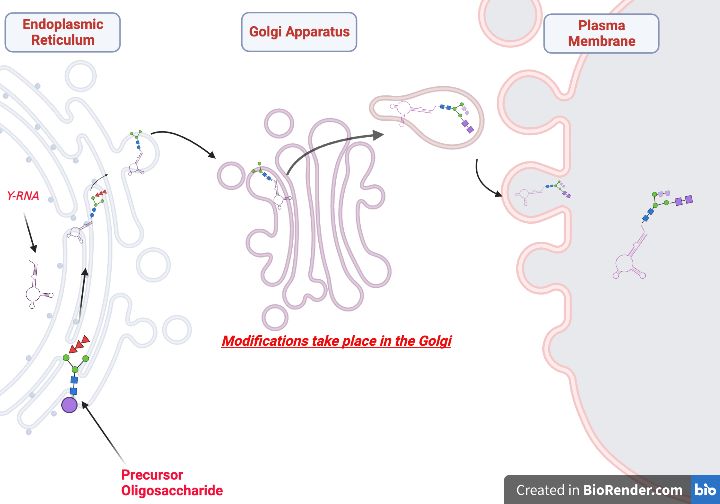 Fig.1 Proposed N-linked glycoRNA biosynthesis process.Distributed under CC BY-SA 4.0, from Wiki, without modification.
Fig.1 Proposed N-linked glycoRNA biosynthesis process.Distributed under CC BY-SA 4.0, from Wiki, without modification.
Why Visualize GlycoRNA Interactions?
Understanding how glycoRNAs interact with proteins or cells is critical for:
- Identifying new drug targets.
- Studying cancer progression and immune diseases.
- Developing diagnostic tools.
Traditional methods (e.g., RNA sequencing) lack real-time insights. Our glycoRNA imaging service solves this by combining high-sensitivity probes with advanced microscopy to track interactions live.
Key Technology: FRET Imaging
FRET (Förster Resonance Energy Transfer) is a gold-standard imaging technique for studying molecular interactions within 1–10 nm distances. At Creative Biolabs, we use a dual-probe fluorescence imaging technique to ensure specificity and accuracy.
How It Works
-
Probe Design:
- Carbohydrate-binding oligonucleotide probes: DNA-based molecules designed to bind glycans such as Neu5Ac or GlcNAc.
- RNA Probes: Fluorescently labeled DNA probes targeting unique RNA sequences (e.g., miRNA, tRNA).
-
Signal Generation:
- When glycoRNA binds to its target (e.g., Siglec receptor), both probes converge, triggering energy transfer between donor (e.g., Cy3) and acceptor (e.g., Cy5) fluorophores.
- FRET signals are captured as a ratio of donor-to-acceptor fluorescence, confirming true interactions.
Advantages Over Traditional FRET
| Feature | Dual-Probe Imaging | Standard FRET |
|---|---|---|
| Specificity | Dual targeting (glycan + RNA) reduces false positives. | Single-probe targeting prone to off-site binding. |
| Sensitivity | Detects low-abundance glycoRNAs (≤10 molecules/cell). | Limited by background noise in complex samples. |
| Dynamic Range | Tracks interactions across physiological pH (6.5–7.5) and temperatures. | Requires strict buffer conditions. |
| Applications | Live-cell imaging, sEV analysis, tissue sections. | Mostly fixed-cell or purified protein studies. |
Why It Matters for GlycoRNA
- Minimized Cross-Reactivity: Probes are pre-validated to avoid binding non-glycosylated RNA or free glycans.
- Quantitative Data: FRET efficiency (%) measures interaction strength, useful for comparing diseased vs. healthy samples.
- Adaptability: Compatible with 3D cell cultures and patient-derived samples for translational research.
Workflow of GlycoRNA Imaging Service
Our step-by-step workflow ensures precise, reproducible results:
1. Sample Preparation
-
Metabolic Labeling:
- Cells/tissues are treated with azide-modified sialic acid precursors (e.g., Ac4ManNAz) for 48–72 hours. This labels glycoRNA glycans with bioorthogonal tags.
- Optional: Click chemistry (e.g., DBCO-Cy5) can further tag glycans for multi-color imaging.
-
Quality Control:
- Flow cytometry validates glycan labeling efficiency (>90% required).
- RNase-free conditions prevent RNA degradation.
2. Probe Binding
-
Probe Optimization:
- GRPs and RNA probes are titrated to achieve optimal signal-to-noise ratios.
-
Incubation:
- Probes are added to live or fixed cells for 1–2 hours.
- Unbound probes are washed away using low-salt buffers.
3. Imaging
-
Microscopy Setup:
- Confocal Microscopy: Captures high-resolution 3D images of glycoRNA distribution (e.g., cell surface vs. cytoplasm).
- TIRF Microscopy: Visualizes glycoRNA interactions at the cell membrane with <100 nm depth penetration.
-
Data Acquisition:
- Dual-channel imaging (donor: 568 nm, acceptor: 647 nm) with time-lapse capabilities.
- FRET signals are quantified using ratiometric analysis.
4. Analysis & Reporting
-
Interaction Validation:
- Co-localization maps confirm glycoRNA-receptor binding.
- Negative controls (e.g., RNase-treated samples) rule out non-specific signals.
-
Deliverables:
- Raw images + processed data (FRET efficiency, binding kinetics).
- Custom reports with biological interpretation.
Sample Requirements
To ensure optimal results from our glycoRNA imaging service, please adhere to the following sample preparation and submission guidelines:
| Sample Type | Minimum Amount | Preparation Guidelines |
|---|---|---|
| Cell Lines | 1×10⁶ cells | Cultured in appropriate media, ≥90% viability. Avoid contamination. |
| Tissues | 50 mg | Flash-frozen in liquid nitrogen. Avoid RNase contamination. |
| Biofluids (serum, plasma) | 200 µL | Collected in EDTA tubes. Centrifuge at 2,000×g for 10 min before submission. |
| Extracellular Vesicles (sEVs) | 1×10¹⁰ particles | Isolated via ultracentrifugation or commercial kits. Provide purity data if possible. |
Please Pay Attention to:
- Labeling: Pre-label samples with unique IDs and include metadata (e.g., cell type, treatment conditions).
- Storage: Cells/Tissues are required to be stored at -80°C or in liquid nitrogen. Biofluids/sEVs are stable at -80°C, please avoid repeated freeze-thaw cycles.
- Shipping: Use dry ice for tissues/sEVs; ice packs for cells/biofluids.
Benefits of Our Service
- Compatible with a range of biological materials.
- Track interactions in live cells.
- Detect low-abundance glycoRNAs (sample volume ≥10 µL).
- Customized protocols and data analysis (PCA, ROC curves).
- Suitable for cancer, immunology, and drug development.
- Turnkey solutions with 72-hour turnaround for standard projects.
Applications
- Detect cancer-specific glycoRNAs on small extracellular vesicles (sEVs).
- Our service achieves high accuracy in distinguishing cancer vs. non-cancer samples.
- Visualize glycoRNA binding to receptors like Siglec-5, a key player in inflammation.
- Study how glycoRNAs recruit immune cells (e.g., monocytes) to disease sites.
GlycoRNA interactions drive critical biological processes, yet their study demands innovative tools. At Creative Biolabs, our glycoRNA imaging service provides unmatched clarity and precision, enabling breakthroughs in disease research. Contact us to explore how our imaging services can advance your glycoRNA projects. Let's unlock the secrets of cellular communication together.
References:
- Ren, Tingju, et al. "FRET imaging of glycoRNA on small extracellular vesicles enabling sensitive cancer diagnostics." Nature Communications 16.1 (2025): 3391. https://doi.org/10.1038/s41467-025-58490-2
- Flynn, Ryan A., et al. "Small RNAs are modified with N-glycans and displayed on the surface of living cells." Cell 184.12 (2021): 3109-3124. https://doi.org/10.1016/j.cell.2021.04.023
- Zhang, Ningning, et al. "Cell surface RNAs control neutrophil recruitment." Cell 187.4 (2024): 846-860. https://doi.org/10.1016/j.cell.2023.12.033
- Ma, Yuan, et al. "Spatial imaging of glycoRNA in single cells with ARPLA." Nature biotechnology 42.4 (2024): 608-616. https://doi.org/10.1038/s41587-023-01801-z