Novel GlycoRNA Imaging: Visualizing GlycoRNAs in Single Cells
GlycoRNA Background
GlycoRNAs represent a groundbreaking discovery, bridging the worlds of RNA biology and glycobiology. First discovered by Flynn, glycoRNA are found to be small non-coding RNAs, including Y RNAs, small nuclear RNAs (snRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), and transfer RNAs (tRNAs), can be modified with N-linked glycans rich in sialic acid and fucose. Remarkably, glycoRNAs localize predominantly to the cell surface, interacting with Siglec family receptors, particularly Siglec-5, thereby orchestrating critical processes such as immune regulation, cell-cell communication, and signal transduction. With several years of expertise in glycoRNA imaging, Creative Biolabs offers a comprehensive overview of novel glycoRNA imaging methods to gain insights into mapping glycoRNA distributions at the single-cell level.
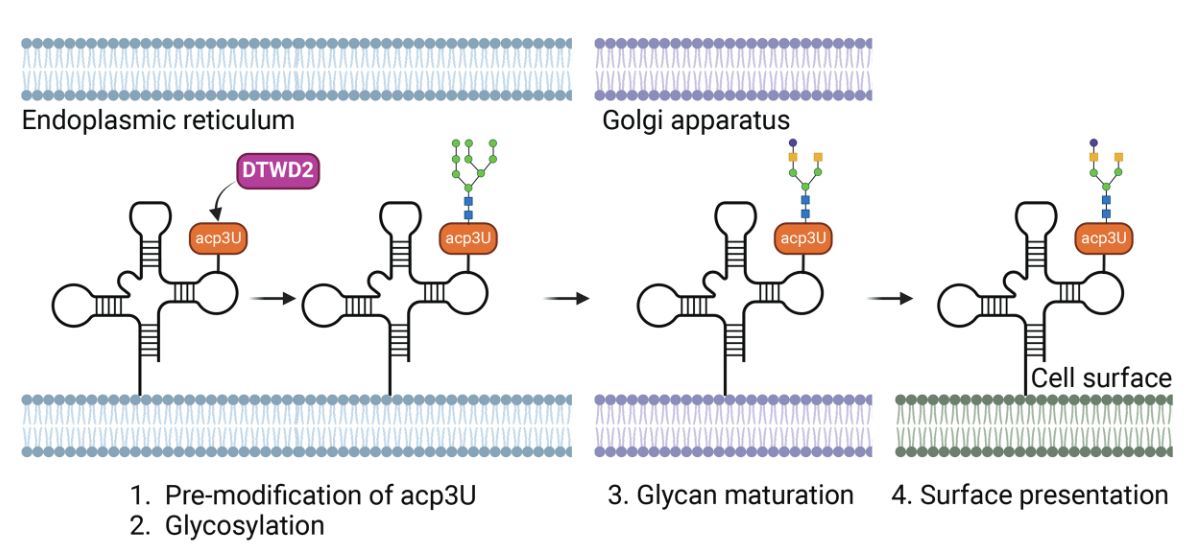 Fig.1 GlycoRNA biosynthesis and surface localization.1
Fig.1 GlycoRNA biosynthesis and surface localization.1
| Feature | Details |
|---|---|
| Composition | Small noncoding RNAs (e.g., Y-RNAs) conjugated with highly sialylated and fucosylated N-glycans |
| Biosynthesis | Dependent on classical N-glycosylation pathways; involves DTWD2-mediated acp3U modification and OST-mediated glycan attachment |
| Localization | Primarily displayed on the external plasma membrane of living cells |
| Functional Roles | Immune modulation, cell-cell signaling, and potential involvement in diseases like cancer and autoimmunity |
Why Detecting GlycoRNAs Matters
Given their central roles in immunity and disease, detecting and characterizing glycoRNAs is crucial. Traditional approaches—such as metabolic labeling combined with mass spectrometry or next-generation sequencing—provide partial views, often missing critical spatial context. Current challenges include:
- Complex, multi-step workflows
- Harsh conditions incompatible with preserving RNA integrity
- Limited ability to simultaneously acquire sequence and spatial information
Thus, an innovative method allowing direct, sensitive, and spatially resolved glycoRNA detection became an urgent need.
Innovative Imaging Approach for GlycoRNAs
A novel imaging method has been developed to address these challenges. This approach integrates nucleic acid binding elements with spatially dependent ligation and signal amplification techniques. The process involves:
-
Dual Recognition:
- A sialic acid-specific aptamer binds the glycan moiety.
- An RNA-specific probe hybridizes to a specific sequence on the glycoRNA.
-
Proximity-Triggered Circularization:
- Spatially adjacent aptamer and RNA probes catalyze the formation of a closed circular DNA template.
-
Signal Amplification:
- The circular DNA template undergoes amplification, significantly enhancing the detection signal.
-
Fluorescent Readout:
- Fluorescently labeled probes hybridize to amplified products, visualized via confocal or super-resolution microscopy.
Novel GlycoRNA Imaging: Sample Compatibility
| Sample Type | Characteristics |
|---|---|
| Fixed Mammalian Cells | HeLa, PANC-1, HEK293T and more. |
| Tumor Cells | Breast cancer models, inflammatory models (e.g., LPS-stimulated cells) |
| Tissue Sections | Both human and animal tissue samples. |
| Subcellular Structures | Lipid rafts, SNARE vesicles |
- No requirement for cell permeabilization.
- Native conformation of glycoRNAs preserved.
- Adaptable to various inflammation and immune activation models.
Novel GlycoRNA Imaging Applications
-
Single-Cell GlycoRNA Mapping
- Direct visualization of glycoRNA subcellular distributions.
- Co-localization with membrane microdomains.
-
Cancer Research
- In breast cancer models, glycoRNA levels inversely correlate with malignancy.
- Potential diagnostic biomarkers.
-
Immune Cell Biology
- GlycoRNAs modulate immune cell adhesion and signaling.
- Dynamic changes during immune activation captured.
-
Intracellular Trafficking Studies
- Tracking glycoRNA exocytosis via SNARE vesicles.
-
Glycobiology Tool Expansion
- Platform adaptable for imaging other glycoconjugates (e.g., glycoproteins).
FRET Imaging vs Novel GlycoRNA Imaging
At Creative Biolabs, we are committed to offering cutting-edge, scientifically validated imaging solutions to advance glycoRNA research. While novel glycoRNA imaging method represents a significant step forward in static visualization of glycoRNAs through ligation-based signal amplification, our FRET-based glycoRNA imaging platform offers distinct advantages in real-time, dynamic molecular interrogation, making it the preferred choice for researchers seeking speed, specificity, and quantitative rigor.
| Feature | Novel GlycoRNA Imaging | FRET Imaging |
|---|---|---|
| Molecular Interaction Resolution | Indirect signal | Direct real-time signal through energy transfer (1–10 nm resolution) |
| Probe Targeting Strategy | Aptamer for glycan + RNA probe (ligation-based) | Simultaneous dual-probe hybridization targeting glycan & RNA (non-enzymatic) |
| Specificity | High (due to dual binding and circular DNA-based signal enhancement) | Very high; dual-recognition minimizes false positives and off-target binding |
| Sensitivity | Single-molecule level via circular DNA-based signal enhancement | Detects ≤10 glycoRNAs/cell without enzymatic amplification |
| Temporal Resolution | Static (end-point detection) | Dynamic; time-lapse FRET imaging supports real-time tracking |
| Workflow Complexity | Multi-step (aptamer binding → ligation → signal enhancement) | Streamlined: direct hybridization and fluorescence acquisition |
| Versatility Across Applications | Optimized for static spatial mapping | Broad: cancer, immunology, inflammation, EV profiling |
Recent Research Progress in GlycoRNA Imaging
A team from Northeastern University developed a dual-recognition Förster resonance energy transfer technique to detect glycoRNA on small extracellular vesicles (sEVs). This method uses nucleic acid probes to detect RNA modified by N-acetylneuraminic acid, enabling sensitive and selective analysis of glycoRNA in minimal biological fluids. The drFRET strategy identified five prevalent sEV glycoRNAs from seven cancer cell lines. In a cohort of 100 patients (covering six types of cancer and non-cancer controls), sEV glycoRNA profiling achieved 100% accuracy in distinguishing cancer cases from non-cancer cases (95% confidence interval) and 89% accuracy in classifying specific cancer types. The study also revealed that sEV glycoRNAs specifically interact with Siglec proteins and P-selectin, which is crucial for sEV internalization. This drFRET approach provides a versatile and sensitive platform for imaging and functional analysis of sEV glycoRNAs, holding great promise for clinical applications such as early cancer screening and precise typing.
These imaging approaches enable high-sensitivity, high-specificity spatial visualization of glycoRNAs at the single-cell level. Our FRET-based system provides quantitative measurements of interaction strength through FRET efficiency analysis. This supports direct comparison between disease states, allowing deeper insight into disease-associated molecular mechanisms. Creative Biolabs remains committed to advancing glycoRNA imaging technologies. Partner with us to push the frontiers of RNA glycobiology.
FAQs
References:
- KIM HS, et al. "GlycoRNA: A new player in cellular communication." Oncol Res. (2025): 995–1000. Distributed under Open Access license CC BY 4.0, without modification. https://doi.org/10.32604/or.2025.060616
- Ma, Yuan, et al. "Spatial imaging of glycoRNA in single cells with ARPLA." Nature biotechnology 42.4 (2024): 608-616. https://doi.org/10.1038/s41587-023-01801-z
- Guo, Weijie, et al. "Sialic acid aptamer and RNA in situ hybridization-mediated proximity ligation assay for spatial imaging of glycoRNAs in single cells." Nature Protocols (2025): 1-21. https://doi.org/10.1038/s41596-024-01103-x