Different Microarrays for Exploring Bacterial Surface Glycans
Finding Your Perfect Match: 4 Microarray Technologies for Bacterial Glycan Research
At Creative Biolabs, we regularly help researchers navigate the microarray landscape. Based on hundreds of projects, here's our straightforward comparison of four powerful platforms for studying bacterial surface glycans. Each has distinct strengths—let's match your goals with the ideal technology.
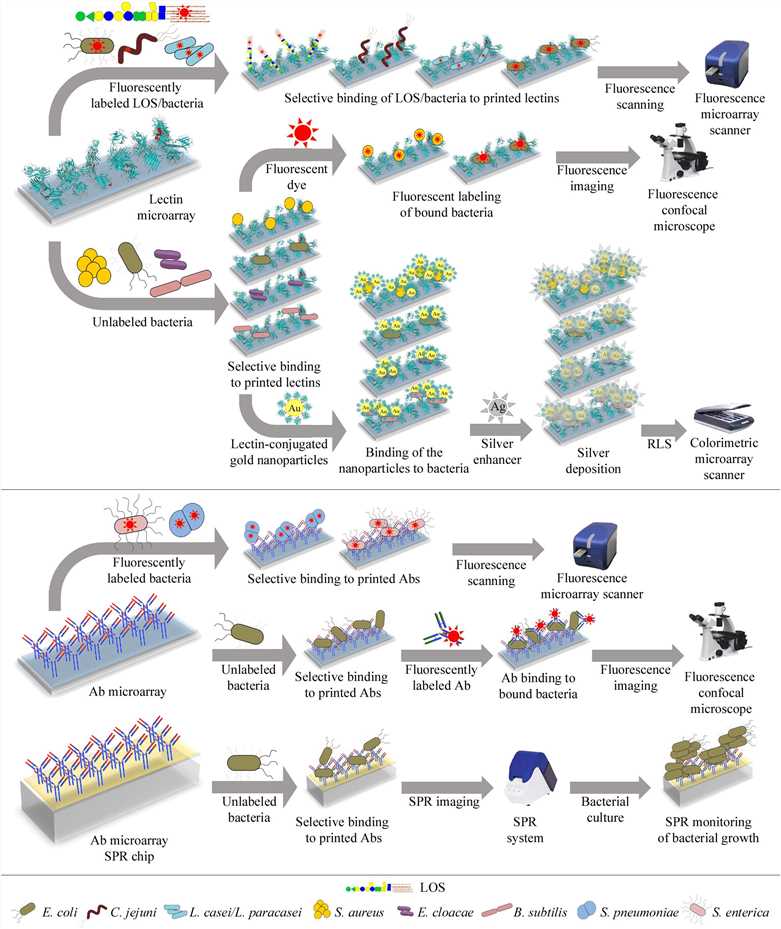 Fig.1 Bacterial detection: Technical pathways of lectin and antibody microarrays.1
Fig.1 Bacterial detection: Technical pathways of lectin and antibody microarrays.1
Need to track specific bacteria or diagnose infections? Our antibody arrays deliver precision. We immobilize custom antibodies that capture target glycans like O-antigens or capsules. Samples are processed rapidly—often detecting pathogens directly in food or clinical specimens. Clients choose this for:
- Outbreak tracing (e.g., identifying E. coli O157 in contaminated beef)
- Vaccine response tracking (measuring antibody levels post-immunization)
Best when you already have specific bacterial targets in mind.
Studying unknown surface sugars? Our lectin arrays excel here. Plant-derived lectins (like ConA or WGA) bind diverse glycans, creating unique "fingerprints" for each bacterium. We've used this to:
- Distinguish Lactobacillus strains missed by genetic testing
- Spot temperature-driven glycan changes in Campylobacter
Choose this for the discovery work on uncharacterized bacteria or dynamic glycan shifts.
Whole Cell Microarrays
Want to see how glycans behave in real life? We print intact bacteria or cell wall fragments. This preserves natural glycan density and 3D arrangement—critical for host-pathogen studies. Recent projects revealed:
- How immune lectins bind differently to isolated glycans vs. live bacteria
- Strain-specific variations in antibiotic penetration
Ideal for teams studying host receptor interactions or antibiotic resistance mechanisms.
Microbial Glycan Arrays (MGMs)
Developing anti-bacterial vaccines? Our MGM platform screens hundreds of purified glycans (LPS, CPS, etc.) against immune proteins. Clients get:
- Rapid antigen candidate screening
- Autoimmune risk assessment (like C. jejuni ganglioside mimics)
We recommend this for glycoconjugate vaccine development and immune escape studies.
Your Decision Checklist
Still unsure? Answer these questions:
- Target known? → Antibody arrays
- Glycans unknown? → Lectin arrays
- Need natural context? → Whole cell arrays
- Vaccine/diagnostics focus? → MGMs
Our teams customize slide chemistries, detection methods, and analysis pipelines for each project type. Let's discuss your bacterial system—we'll propose the most efficient array strategy to accelerate your results.
| Technology | Best For | Key Strengths | Limitations |
|---|---|---|---|
| Antibody Microarrays |
|
|
|
| Lectin Microarrays |
|
|
|
| Whole Cell Microarrays |
|
|
|
| Microbial Glycan Arrays (MGMs) |
|
|
|
Different Microarrays: Illuminating Pathogen-Host Interactions
Decoding Molecular Mimicry in Campylobacter jejuni
Microbial glycan microarrays have transformed glycobiology research by enabling high-throughput analysis of carbohydrate-mediated interactions. This platform immobilizes hundreds of microbial glycans—including lipopolysaccharides (LPS), capsular polysaccharides (CPS), and lipooligosaccharides (LOS)—on microscopic slides. When exposed to immune proteins (antibodies, lectins) or whole pathogens, binding events are detected via fluorescence or surface plasmon resonance. Crucially, this method requires minimal sample volumes while screening entire glycome repertoires, a feat impossible with traditional assays.
The real-world impact of MGMs shines in studies of C. jejuni1, a leading cause of foodborne neuropathy. Researchers discovered that certain strains express LOS structures mimicking human ganglioside GM1—a self-antigen in neural tissues. Using custom lectin microarrays, Semchenko et al. compared LOS from three clinical strains:
- 11168-O (GM1 mimic)
- 81-176 (GM2 mimic)
- 224 (unknown type)
Surprisingly, despite predictions, cholera toxin B (GM1 binder) failed to recognize strain 11168-O. Instead, Gal-specific lectins (PNA, VAA, jacalin) showed intense binding to 11168-O LOS—consistent with terminal galactose in GM1—while strain 81-176 exhibited weaker signals. Even more intriguingly, strain 224 bound anti-GM1 antibodies but not Gal-lectins, suggesting cryptic structural variations. Follow-up work with 15-lectin arrays revealed that PCR-based LOS genotyping often mispredicts actual glycan structures, highlighting MGMs' value in identifying autoimmune triggers.
E. coli O157:H7: Taming a Shape-Shifting Pathogen
For Shiga-toxin-producing E. coli (STEC), MGMs tackle a critical challenge: antigenic variability. Studies1 confirm that O157:H7's O-antigen exhibits striking structural plasticity across strains, complicating vaccine design. Hegde et al. deployed antibody microarrays printed with:
- Anti-O-antigen antibodies (O26, O45, O103, etc.)
- Corresponding purified O-polysaccharides
When tested against enriched ground beef samples contaminated with 1–10 CFU/g, the arrays specifically detected all six major non-O157 STEC serogroups without cross-reactivity. But here's the breakthrough: SPRi-based microarrays eliminated pre-enrichment needs by monitoring real-time bacterial growth on antibody-functionalized chips, detecting ≤5 CFU/g—a sensitivity leap for food safety.
Klebsiella pneumoniae: Capsule Complexity Unmasked
K. pneumoniae's capsule—a critical virulence shield—became far less enigmatic through MGM studies. Strain 52145's surface architecture features both O-antigen (Gal-rich) and CPS (Glc/Man-based)1. Lectin microarray profiling revealed:
- RCA/PNA lectins bound primarily to O-chain Gal residues (not CPS)
- ConA recognized non-CPS Man/Glc epitopes
Capsule removal enhanced O-antigen accessibility. This explained why antibodies struggled to target encapsulated strains. MGMs further identified the conserved LPS inner core tetrasaccharide [GlcNAcα(1-2)Hepα(1-3)Hepα(1-5)Kdo] as a vaccine candidate. When conjugated to CRM197 carrier protein, this glycan induced opsonic antibodies in mice—a pivotal step toward fighting carbapenem-resistant strains.
Practical Innovations
MGMs are catalysts for engineering solutions:
- Antibody Discovery: Microarrays identified two monoclonal antibodies (mAbs) that killed drug-resistant K. pneumoniae by targeting CPS epitopes.
- Phage Therapy: Gp047—a bacteriophage protein—was shown to bind acetamidino-modified pseudaminic acid on C. jejuni flagella, inspiring motility-disrupting therapies.
- Plant-Bacteria Symbiosis: LysM domain microarrays revealed how Rhizobium's lipochitin oligosaccharides engage plant receptors, suggesting bioengineering applications.
Despite successes, limitations persist. Some lectins show inconsistent binding due to immobilization methods. Expanding glycan libraries remains arduous—though "glycophage" displays offer promise. Crucially, whole-bacteria microarrays now complement purified glycan studies, capturing native surface architecture. As synthetic chemistry advances, MGMs will accelerate precision vaccines against ever-evolving pathogens. At Creative Biolabs, we help you choose the right microarray tool to crack your glycan code. Contact us for more microarray selection tips and service details.
Reference:
- Campanero-Rhodes, María Asunción, et al. "Microarray strategies for exploring bacterial surface glycans and their interactions with glycan-binding proteins." Frontiers in Microbiology 10 (2020): 2909. Distributed under Open Access license CC BY 4.0 , without modification. https://doi.org/10.3389/fmicb.2019.02909
