U2AF1 ensures precise exon-intron discrimination, a process critical for generating protein diversity. Mutations disrupt this fidelity, leading to isoforms with altered function.
Introduction to U2AF1
U2AF1 (U2 small nuclear RNA auxiliary factor 1) is a critical component of the spliceosome, the molecular machinery responsible for pre-mRNA splicing. As a subunit of the U2AF heterodimer, U2AF1 recognizes the 3' splice site AG dinucleotide, ensuring precise excision of introns and ligation of exons. Mutations in U2AF1, particularly in its conserved zinc-finger domains, are linked to myeloid malignancies like myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), where they disrupt normal splicing patterns, leading to aberrant expression of oncogenes and tumor suppressors.
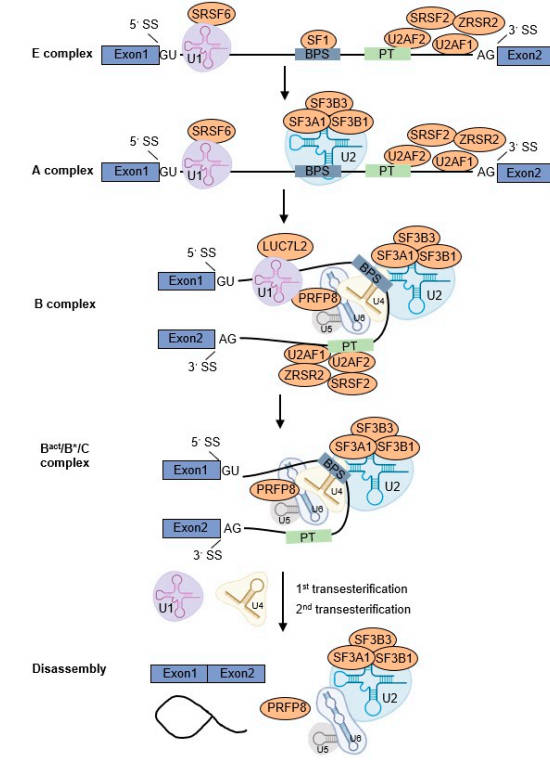 Fig.1 U2AF1 participates in the pre-mRNA splicing process.1
Fig.1 U2AF1 participates in the pre-mRNA splicing process.1
Biological Roles
Splicing Regulation
Hematopoietic Homeostasis
In myeloid cells, U2AF1 mutations drive clonal expansion by mis-splicing genes involved in differentiation. This contributes to myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).
Stress Response
U2AF1 modulates alternative splicing of heat shock proteins (HSPs) and unfolded protein response (UPR) genes, enabling cells to adapt to proteotoxic stress. Dysregulation here links U2AF1 to neurodegenerative diseases.
Tumor Microenvironment
Aberrant splicing of cytokines and adhesion molecules alters immune cell infiltration and angiogenesis, fostering a pro-tumorigenic niche.
Cutting-Edge Detection Platforms
Whole-Exome Sequencing (WES)
This gold-standard approach enables comprehensive profiling of somatic mutations in U2AF1 and its interacting splicing factor genes. By targeting coding regions, WES efficiently identifies pathogenic variants linked to myeloid malignancies and solid tumors.
RNA-Seq
A transformative tool for quantifying splicing dysregulation, RNA-Seq captures global changes in transcriptome architecture. It resolves exon skipping events, intron retention, and alternative 3' splice site usage, which may activate cryptic exons or truncate key proteins.
Western Blotting (WB)
Paired with differential expression analysis, WB offers precise quantification of U2AF1 protein levels across cellular and tissue samples. Using high-affinity rabbit polyclonal antibodies, this technique distinguishes wild-type U2AF1 from mutant isoforms that may exhibit altered stability or localization. WB is indispensable for validating RNA-level findings and correlating protein abundance with phenotypic outcomes.
Immunoprecipitation (IP)
IP isolates U2AF1-containing protein complexes to dissect its interactions with splicing enhancers or co-factors like U2AF2. By coupling IP with mass spectrometry (IP-MS), researchers can map U2AF1's dynamic interactome under normal or pathological conditions. This approach is particularly valuable for identifying novel splicing regulators or therapeutic targets in U2AF1-driven diseases.
Applications
Mechanistic Studies
Cancer Pathogenesis: U2AF1 mutations disrupt 3' splice site selection, leading to aberrant isoforms of oncogenes and tumor suppressors, driving tumorigenesis in hematological and solid malignancies.
Disease Networks
RNA-Seq and proteomic analyses reveal mutation-specific splicing signatures linked to metabolic reprogramming, DNA repair defects, and therapy resistance.
Biomarker Development
Prognostic Markers: U2AF1 mutation burden correlates with aggressive disease phenotypes and shorter survival in myeloid neoplasms.
Predictive Tools
Splicing profiles predict responses to targeted therapies and immune checkpoint blockade.
Cross-Disease Mechanistic Insights
Rare Genetic Disorders: U2AF1 mutations cause mis-splicing in inherited diseases, informing gene therapy strategies.
Stress Response Pathways
Aberrant U2AF1 activity alters stress granule dynamics, linking splicing defects to cellular adaptation and disease progression.
Drug Design
High-resolution structures of U2AF1-RNA complexes guide rational development of splicing inhibitors.
FAQs
-
What sample types are compatible with your U2AF1 analysis platform?
Our workflow accommodates a wide range of biological specimens, including:
- Fresh or frozen tissue (solid tumors, bone marrow aspirates)
- Formalin-fixed paraffin-embedded (FFPE) sections (archival pathology samples)
- Blood-derived samples (peripheral blood mononuclear cells, PBMCs; plasma for cfDNA analysis)
- Cell lines (primary patient-derived cells, immortalized lines for functional studies)
-
How do U2AF1 mutations affect non-coding RNA splicing?
U2AF1 dysregulation alters:
lncRNA processing: Disrupting splicing of MALAT1 or NEAT1, which regulate nuclear architecture and gene expression.
circRNA biogenesis: Shifting back-splicing efficiency, potentially affecting microRNA sponging or protein binding.
Epigenetic crosstalk: U2AF1-mutant cells show altered DNA methylation patterns linked to splicing factor recruitment.
-
Can U2AF1 mutation status guide immunotherapy responses?
Preliminary data indicate:
- Neoantigen generation: U2AF1-driven splicing errors may create tumor-specific peptides recognized by T cells.
- Checkpoints: U2AF1-mutant tumors upregulate PD-L1 splicing variants, suggesting synergy with PD-1 inhibitors.
- Microbiome links: Gut microbiota may modulate U2AF1-related immune responses via short-chain fatty acids.
Reach out to us to receive a customized quote for U2AF1 analysis!
Reference
- Zhao, Yangjing, et al. "The biological and clinical consequences of RNA splicing factor U2AF1 mutation in myeloid malignancies." Cancers 14.18 (2022): 4406. Distributed under Open Access license CC BY 4.0, without modification. https://doi.org/10.3390/cancers14184406
For Research Use Only.

