Organelle-Targeted Liposome System Design
Liposomes can be engineered not just to enter cells, but to home to specific organelles—mitochondria, lysosomes, endoplasmic reticulum (ER), Golgi, or nucleus—so that bioactive payloads act where biology happens. Creative Biolabs designs organelle-targeted liposome systems that bring intracellular precision to your research.
Trusted by Industry Leaders
Leading biopharma and academic teams rely on Creative Biolabs for formulation science, ligand conjugation, and advanced intracellular targeting analytics.





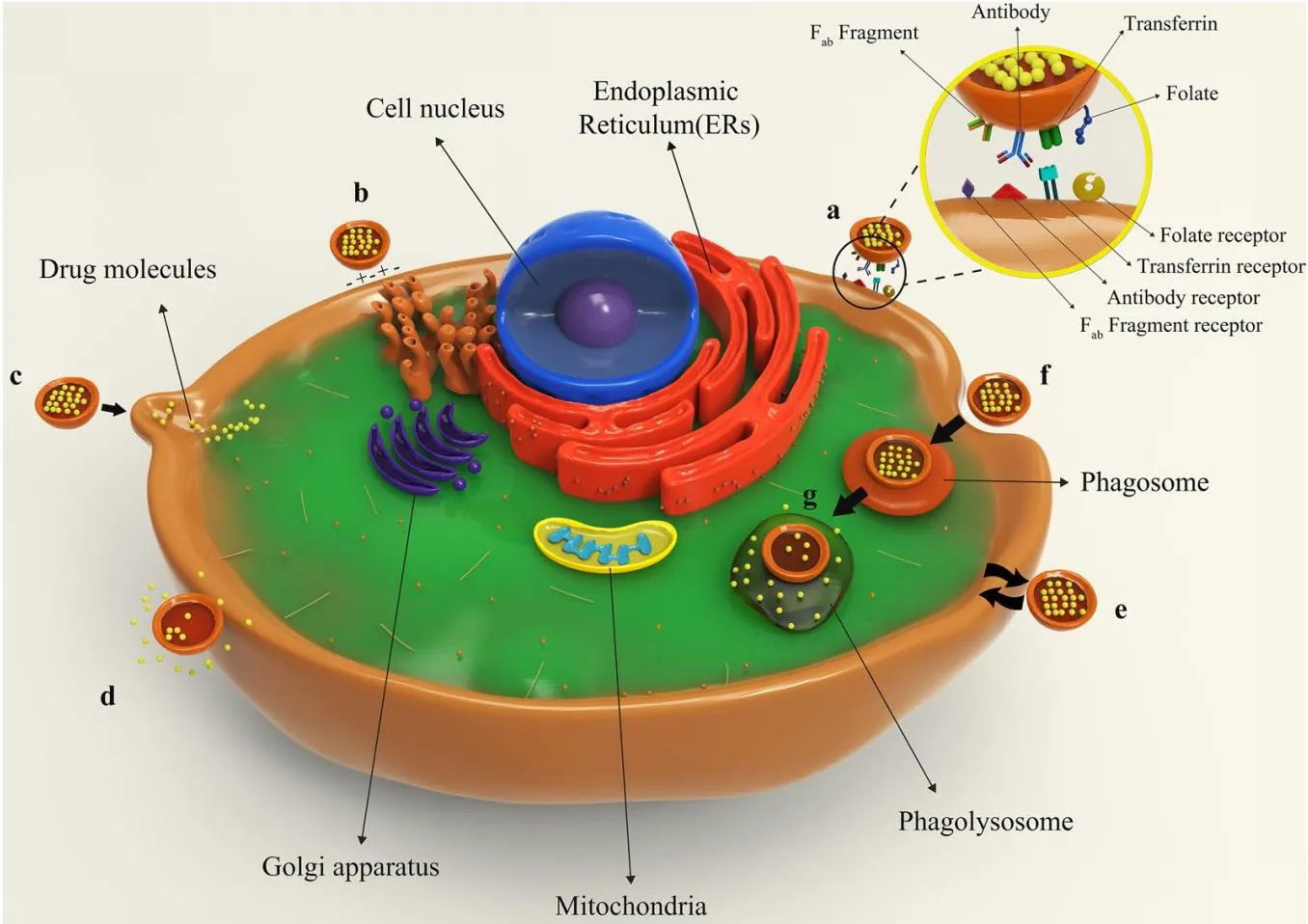
Why Organelle-Targeted Liposome Design Matters
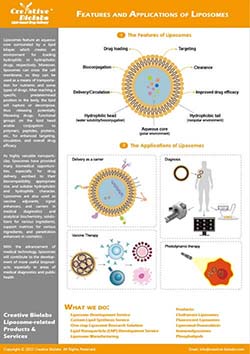
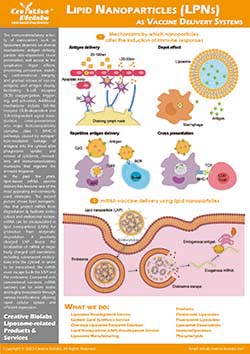
Most intracellular targets reside inside organelles. Delivering payloads to the right compartment improves potency in model systems, reduces off-target effects, and clarifies mechanism. Modern strategies combine organelle-addressing ligands—such as triphenylphosphonium for mitochondria, morpholine or mannose-6-phosphate for lysosomes, and nuclear localization signals for the nucleus—with stimuli-responsive membranes and validated colocalization analytics to confirm where cargo actually goes.
Custom Organelle Targeting Strategies
- Mitochondria: Lipid-anchored cations like triphenylphosphonium accumulate payloads by leveraging the mitochondrial membrane potential.
- Lysosome: Weak-base motifs such as morpholine and mannose-6-phosphate (M6P) conjugates exploit acid trapping or receptor-mediated uptake.
- Nucleus: NLS-bearing peptides guide liposomes through the nuclear pore complex when size and timing are optimized.
- ER/Golgi: ER-retention sequences and small-molecule Golgi probes are adapted into conjugation scaffolds to direct liposomes to these compartments.
Ligands are installed by orthogonal chemistries—maleimide–thiol coupling, copper-free click chemistry (SPAAC), or CuAAC under chelated conditions—balancing conjugation yield, ligand orientation, and membrane integrity.
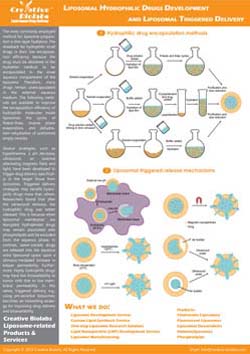
- Stimuli-responsive bilayers: DOPE/CHEMS compositions destabilize in acidic compartments, accelerating release.
- Endosomal escape boosters: Fusogenic peptides and buffering lipopolymers help transfer cargo into the cytosol before organelle entry.
- Cargo matching: Hydrophilic payloads use aqueous core loading, hydrophobic compounds partition into the bilayer, and sensitive macromolecules may require cleavable linkers.
Creative Biolabs doesn’t stop at uptake. We quantify organelle targeting with confocal colocalization (Pearson’s and Manders’ coefficients with thresholding controls) and subcellular fractionation followed by western blotting against canonical markers (e.g., COX IV for mitochondria; LAMP1 for lysosome; calnexin for ER; GM130 for Golgi; histone H3 for nucleus).
Service Scope and Capabilities
| Module | Key Features | Representative Approaches |
|---|---|---|
| Organelle-Specific Targeting Modules | Tailored liposomes designed to accumulate in specific organelles |
|
| Advanced Conjugation & Formulation | Broad bioconjugation chemistry and formulation control |
|
| Quantitative Targeting Analytics | Rigorous validation of organelle localization |
|
Service Workflow
Project Consultation
Define organelle, model, and research goals.
Design Blueprint
Select ligands, lipids, and escape strategies.
Prototype Fabrication
Liposome formulation and ligand installation.
Characterization
DLS, zeta potential, encapsulation efficiency, stability.
Validation
Confocal colocalization, fractionation, and functional assays.
Optimization & Reporting
Refine composition and deliver complete data package.
Deliverables
- Detailed formulation dossier with lipid composition and methods.
- QC report with DLS, PDI, zeta potential, and encapsulation efficiency.
- Targeting evidence including imaging, colocalization analysis, and fractionation data.
- SOPs for ligand conjugation and analytics.
- Consultation summary with optimization insights.
All services are strictly For Research Use Only. Not For Clinical Use.
Advantages of Our Service
Expertise in advanced lipid modification and conjugation chemistries
Broad range of targeting ligands for mitochondria, lysosomes, ER, and nuclei
High reproducibility and scalability from pilot to research-grade production
Integrated analytical platforms for rigorous quality assessment
Flexible customization to meet diverse research needs
Applications of Organelle-Specific Liposomes
Mitochondrial research
Delivery of probes or nucleic acids for respiration, mitophagy, or ROS studies.
Lysosomal biology
Delivery of substrates or reporters to study degradation kinetics and pH dynamics.
ER stress and proteostasis
Target ER with unfolded-protein-response reporters or modulators.
Golgi trafficking
Explore glycosylation and cargo sorting with Golgi-targeted carriers.
Nuclear delivery studies
Introduce ribonucleoproteins or dyes with NLS-assisted entry to assess nuclear import.
Related Services from Creative Biolabs
Beyond organelle-targeted liposome systems, Creative Biolabs offers a broad portfolio of lipid-based delivery solutions to support comprehensive research programs:
Frequently Asked Questions
Organelle-targeted liposomes are engineered nanoparticles modified with ligands like TPP or NLS to deliver agents directly into mitochondria, nucleus, or lysosomes for precise intracellular delivery.
Other Resources
References
- Sanchez-Aranguren, Lissette, et al. "Mitochondrial-targeted liposome-based drug delivery–therapeutic potential and challenges." Journal of Drug Targeting 33.5 (2025): 575-586. https://doi.org/10.1080/1061186X.2024.2437440
- Rommasi, Foad, and Neda Esfandiari. "Liposomal nanomedicine: applications for drug delivery in cancer therapy." Nanoscale research letters 16.1 (2021): 95. https://doi.org/10.1186/s11671-021-03553-8 (Distributed under Open Access license CC BY 4.0, without modification.)
- Yang, Jingjing, et al. "Organelle-targeted therapies: a comprehensive review on system design for enabling precision oncology." Signal transduction and targeted therapy 7.1 (2022): 379. https://doi.org/10.1038/s41392-022-01243-0
- Tang, Wilson, et al. "Design principles and biomedical applications of endoplasmic reticulum-targeting luminescent nanoparticles." Nano Research 18.6 (2024). https://doi.org/10.26599/NR.2025.94907356
Online Inquiry
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.