Glycosylation Site Analysis
Although various glycosylation reactions create a wide diversity of oligosaccharide structures by different glycosylases, N- and O-linked glycans are the most common to all eukaryotes. N- and O-linked glycans are attached to the asparagine residues as well as serine or threonine residues respectively, while not every suitable residue has to be modified with an oligosaccharide chain, which depends on the type and activity of enzymes in the environment. However, the glycosylation site distribution impacts the structure and function of glycoproteins due to the improvement of physicochemical properties and cell recognition bioactivities of oligosaccharide chains. Creative Biolabs is one of the well-recognized experts who are professionals in offering high-quality protein engineering services.
N-Linked Glycosylation Site Analysis
N-linked glycosylation often occurs on the asparagine residues and is featured by conserved pentasaccharide core including three Man and two GlcNAc. The reducing end of N-linked glycan is a GlcNAc which binds to the Asp residue covalently. There are two common ways to measure the site of N-glycosylation. One is incomplete release and the other is deamidation mechanism. The former is using endoglycosidase which releases N-glycans but leaves a GlcNAc core attached to the glycopeptide. By mapping the location of the remaining GlcNAc by LC-MS, we can gain information on the site location as well as the site occupancy. However, due to the limited site-specificity, not all N-glycans are cleaved, making this approach not generally applicable. By far, the most widely accepted way to analyze the site of N-glycosylation is 18O-labeling. Enzymatic deglycosylation converts asparagine residues at the N-glycosylation site of the glycopeptide to aspartic acids via a deamidation mechanism by the addition of an 18O atom from the surrounding heavy water. This increases the mass of the peptide by 2.9882 Da on the N-glycosylation site and thus enables the identification of sites of glycosylation. The location of the 18O-labeled aspartic acid residues is then mapped by LC-MS.
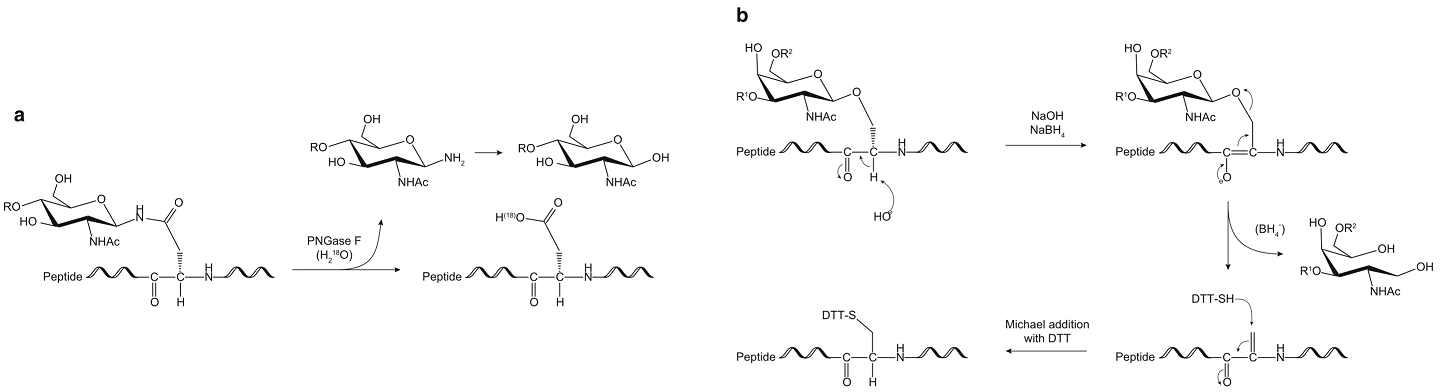 Fig.1 Schematic representation showing the mechanism of N-glycans 18O-labeling (a) and O-glycans by reductive β-elimination and optional BEMAD (b).1, 2
Fig.1 Schematic representation showing the mechanism of N-glycans 18O-labeling (a) and O-glycans by reductive β-elimination and optional BEMAD (b).1, 2
O-Linked Glycosylation Site Analysis
O-linked glycosylation often occurs on the serine or threonine residues and the release is usually accomplished chemically, most commonly by reductive β-elimination. This release of O-glycans leaves behind a modified serine or threonine residue in the former glycopeptide. This residue is reactive toward the thiol functionality, and this property is exploited in the β-elimination by Michael addition with dithiothreitol (BEMAD) method to tag this residue for unambiguous MS detection. The reagent of choice for this purpose is DTT, which introduces a 136.0 Da mass shift compared to non-O-glycosylated serine or threonine.
Glycosylation site analysis ensures the accurate structure determination of specific glycoproteins and uniformity of glycoprotein drug batches. Creative Biolabs provides comprehensive and one-stop glycosylation site analysis services to our clients all over the world. Please feel free to contact us for more details.
References
-
Shajahan, Asif, et al. "Glycomic and glycoproteomic analysis of glycoproteins—a tutorial." Analytical and bioanalytical chemistry 409 (2017): 4483-4505.
-
Under Open Access license CC BY 4.0, without modification.
For Research Use Only.
Resources
 Fig.1 Schematic representation showing the mechanism of N-glycans 18O-labeling (a) and O-glycans by reductive β-elimination and optional BEMAD (b).1, 2
Fig.1 Schematic representation showing the mechanism of N-glycans 18O-labeling (a) and O-glycans by reductive β-elimination and optional BEMAD (b).1, 2