Introduction of Ubiquitylation
Ubiquitylation is a crucial post-translational modification (PTM) process in which ubiquitin, a small protein consisting of 76 amino acids, is covalently linked to specific target proteins. This intricate process, mediated by a cascade of E1, E2, and E3 enzymes, orchestrates diverse cellular functions including protein degradation, signal transduction, DNA repair, and immune responses.
-
Ubiquitin-Activating Enzyme (E1): Activates ubiquitin in an ATP-dependent manner, forming a thioester bond.
-
Ubiquitin-Conjugating Enzyme (E2): Accepts ubiquitin from E1 via a trans-thioesterification reaction.
-
Ubiquitin Ligase (E3): Transfers ubiquitin from E2 to a lysine residue on the target protein, forming an isopeptide bond. E3 ligases are key for substrate specificity.
-
Deubiquitinating enzymes (DUBs): They are a group of proteases responsible for removing ubiquitin from target substrates or cleaving ubiquitin chains, effectively reversing the modification and facilitating the recycling of ubiquitin.
Ubiquitin can be attached as a single molecule (monoubiquitination), multiple single molecules on different lysines (multiubiquitination), or as polyubiquitin chains. Polyubiquitin chains can form via any of the seven lysine residues (K6, K11, K27, K29, K33, K48, K63) within ubiquitin itself, or via its N-terminal methionine. Each linkage type can convey distinct biological signals: K48-linked chains are the canonical signal for proteasomal degradation, while K63-linked chains are typically involved in non-proteolytic functions like DNA repair and signaling. K11-linked ubiquitin chains have been associated with proteasomal degradation, especially in the context of endoplasmic reticulum-associated degradation (ERAD). The complexity is further amplified by mixed-linkage chains and post-translational modifications on ubiquitin itself (e.g., Phosphorylation, Acetylation). The quantitative analysis of ubiquitylated proteins, or the "ubiquitylome", is crucial for deciphering these complex regulatory networks and identifying therapeutic targets in diseases like cancer, neurodegeneration, and infectious diseases. Creative Biolabs utilizes advanced mass spectrometry (MS) techniques and enrichment methods to deliver an in-depth analysis of this essential proteomic landscape.
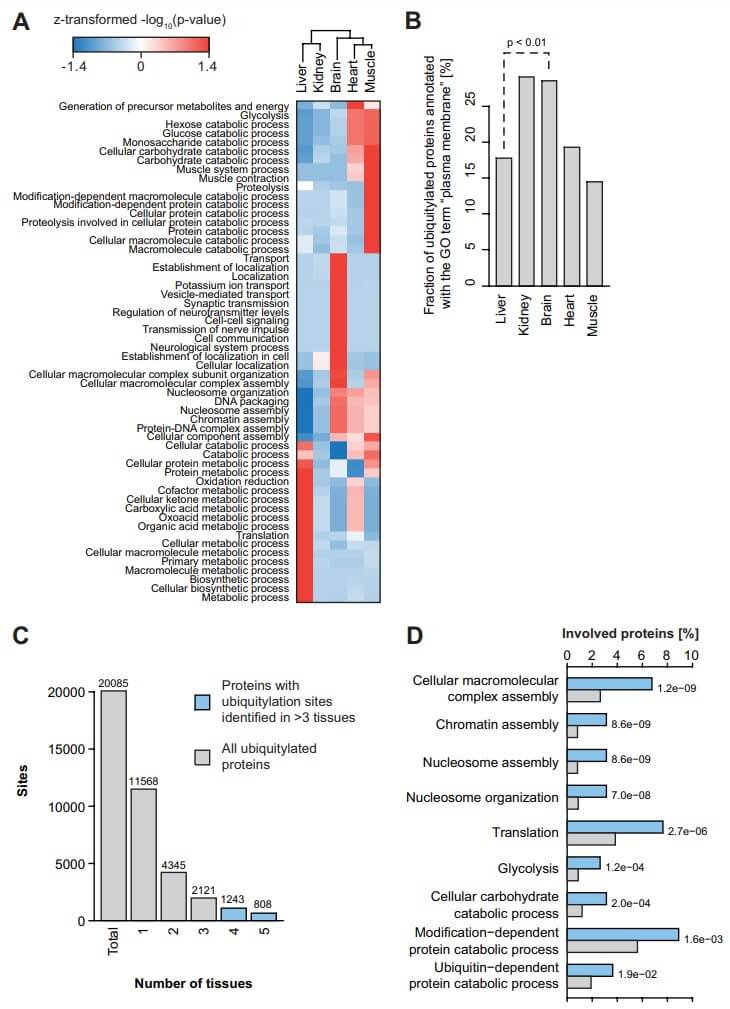 Fig.1 Biological processes associated with ubiquitinated proteins.1
Fig.1 Biological processes associated with ubiquitinated proteins.1
Proteomics Technologies for Quantitative Analysis of Ubiquitinated Proteomes
MS has emerged as the premier technology for large-scale, quantitative ubiquitome analysis. The core principle relies on the fact that trypsin digestion of ubiquitylated proteins leaves a characteristic di-glycine (K-ε-GG) remnant attached to the modified lysine residue. This di-glycine "footprint" serves as a unique signature for ubiquitylation. Key quantitative analysis technologies include:
Stable isotope labeling by amino acids in cell culture (SILAC)
Cells are grown in media containing "light" or "heavy" (isotope-labeled) amino acids. Samples are mixed before processing, allowing for precise relative quantification of ubiquitylation changes.
Tandem mass tag (TMT) / isobaric tags for relative and absolute quantitation (iTRAQ)
Chemical tags with distinct isotopic masses are used to label peptides from different samples. These tags become isobaric in the MS1 scan but yield unique reporter ions upon fragmentation (MS2/MS3), enabling multiplexed quantification of up to 10 or more samples in a single MS run.

Label-free quantification (LFQ)
Relies on comparing peptide ion intensities or spectral counts across different MS runs without isotopic labels. While offering high throughput, it can be more susceptible to run-to-run variability.
High-resolution mass spectrometry (LC-MS/MS)
Advanced instruments like Q-Exactive are coupled with nano-liquid chromatography for sensitive and accurate peptide separation and detection. Fragmentation techniques like higher-energy collisional dissociation (HCD) and synchronous precursor selection MS3 (SPS-MS3) are crucial for obtaining high-quality fragment ion spectra, essential for confident site localization and quantification.
Ubiquitination Modification Proteomic Quantitative Analysis Service: Accelerate Your Drug Discovery Process!
Our meticulously designed workflow ensures comprehensive and reliable analysis, providing you with actionable data. Our service encompasses several critical stages, each optimized for maximum data quality and biological relevance.
-
Sample Preparation and Lysis
This initial step is crucial for preserving the integrity of ubiquitylation. Cells or tissues are lysed under denaturing conditions (e.g., using 8M urea, strong detergents) to immediately halt enzymatic activity and prevent deubiquitination. Key inhibitors like N-ethylmaleimide (NEM), chloroacetamide, and a protease inhibitor cocktail are added to maintain the ubiquitylation state. Proteins are then quantified accurately (e.g., via BCA assay), reduced, and alkylated to ensure consistent downstream processing.
The prepared proteins are precisely digested into smaller peptides. This is typically a two-step enzymatic process, starting with Lys-C followed by trypsin. This digestion specifically cleaves proteins at lysine and arginine residues, generating characteristic di-glycine (K-ε-GG) remnants at ubiquitylated lysine sites. These remnants are the unique "footprints" used for identification.
-
Ubiquitylated Peptide Enrichment
This is a cornerstone of Creative Biolabs's service, addressing the low abundance of ubiquitylated peptides. We employ highly specific anti-di-glycine remnant antibodies coupled with advanced immunoaffinity purification techniques. This step efficiently isolates ubiquitylated peptides from the complex peptide mixture.
For quantitative comparisons across multiple samples, we can label enriched peptides with isobaric tags. This allows peptides from different experimental conditions to be combined into a single mixture. This approach significantly improves throughput, reduces instrument time, and minimizes technical variability across replicates.
-
High-Resolution Mass Spectrometry
The labeled or label-free peptides are separated by high-performance liquid chromatography (HPLC), which reduces sample complexity and improves detection. The separated peptides are then analyzed using mass spectrometers. These instruments provide high mass accuracy and resolution, crucial for confident identification and quantification of ubiquitylation sites.
Raw MS data is meticulously processed using sophisticated software for peptide and protein identification, ubiquitylation site localization, and quantitative analysis. Comprehensive bioinformatics tools are then applied to conduct rigorous statistical analysis, functional annotation, and hierarchical clustering of ubiquitylation patterns, thereby transforming complex data into meaningful biological insights.
Importance and Application of Ubiquitination Modification Proteomic Analysis
Biomarker discovery
Identifying ubiquitylation-based biomarkers for disease diagnosis, prognosis, and therapeutic response monitoring.
Drug target validation
Confirming the mechanism of action of drugs targeting (e.g., proteasome inhibitors, E3 ligase modulators) and identifying off-target effects.
Mechanistic studies
Elucidating the specific roles of different ubiquitin chain linkages (K48, K63, K11, etc.) in various cellular processes.
Understanding host-pathogen interactions
Revealing how pathogens manipulate the host ubiquitylation machinery for their replication and immune evasion.
Fundamental biology
Expanding the known ubiquitylome, identifying novel substrates, and understanding the interplay between ubiquitylation and other PTMs (e.g., phosphorylation, SUMOylation).
Creative Biolabs is your trusted partner for advanced ubiquitylation proteomics, committed to delivering high-quality, actionable data that drives your scientific discoveries. Our comprehensive service, backed by cutting-edge technology and expert analysis, empowers you to unravel the complexities of protein regulation and accelerate your research and drug development initiatives. In addition, we also provide a variety of Glycoproteomics Quantitative Analysis services, such as N-Glycan-based Glycoproteomic and O-Glycan-based Glycoproteomic Analysis. Please contact our team to discuss your specific research needs, and we will provide a tailored solution.
Published Data
This research delves into the intricate patterns of protein ubiquitylation across various mouse tissues, providing a comprehensive map of these crucial post-translational modifications. The study employed a sophisticated workflow that combined high-resolution mass spectrometry with a specialized technique for isolating di-glycine-modified peptides, which are telltale signs of ubiquitylation. Researchers meticulously prepared samples from the brain, heart, kidney, liver, and skeletal muscle. These tissues underwent digestion, and the resulting peptides were enriched using two distinct di-glycine-specific antibodies before being analyzed by advanced mass spectrometers. The findings, visually represented in the Figure below, reveal the scale and distribution of ubiquitylation. Over 20,000 unique ubiquitylation sites were pinpointed, marking the largest dataset of its kind in mammalian tissues to date. Notably, the liver exhibited the highest number of identified sites, suggesting its highly dynamic protein turnover. In contrast, skeletal muscle yielded the fewest sites. A significant portion, roughly 58% of these sites, were found to be tissue-specific, with the brain contributing a particularly large number of unique modifications, underscoring its unique biological complexity. To ensure data robustness, the study included multiple replicate experiments for each tissue, demonstrating high reproducibility, with over 70% of the reported sites consistently identified across different runs. For instance, liver experiments, conducted in triplicate, showed substantial overlap, ranging from 67% to 92% of identified sites.
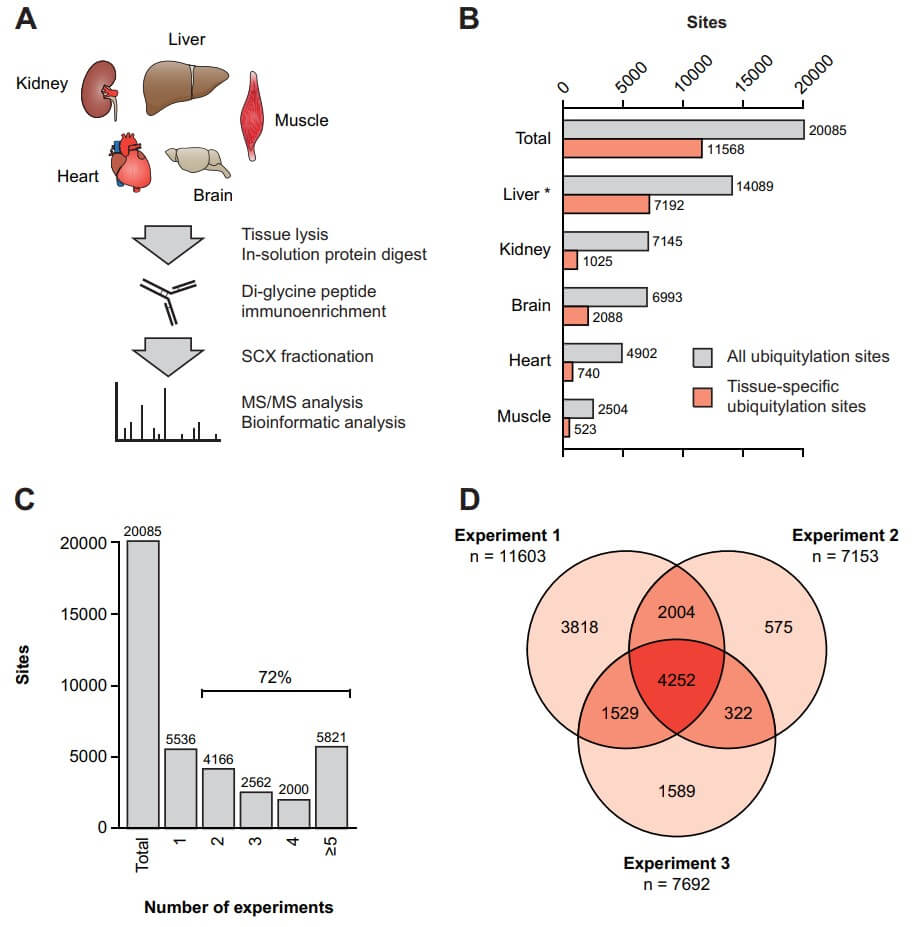 Fig.2 A protocol for the analysis of endogenous ubiquitination sites in mouse tissues using MS.1
Fig.2 A protocol for the analysis of endogenous ubiquitination sites in mouse tissues using MS.1
FAQs
Q1: What is the typical turnaround time for a ubiquitylation proteomics project?
A1: The typical timeframe ranges from 6 to 12 weeks, depending on the complexity of your samples and the scope of the analysis. We prioritize efficiency without compromising data quality. For a more precise estimate tailored to your specific needs, we encourage you to contact our team for a consultation.
Q2: What kind of bioinformatics support do you provide after the MS analysis?
A2: We provide extensive bioinformatics support, including rigorous statistical analysis, Gene Ontology (GO) enrichment, pathway analysis, and hierarchical clustering. Our goal is to transform raw data into clear, interpretable biological insights, helping you understand the functional implications of ubiquitylation changes in your system.
Q3: How does Creative Biolabs's service compare to traditional methods for studying ubiquitylation?
A3: Our service offers significant advantages over traditional methods like Western blotting or single-site analyses. We provide global, site-specific, and quantitative insights into the ubiquitylome, enabling the discovery of novel ubiquitylation events and the comprehensive mapping of dynamic changes across thousands of proteins simultaneously. This high-throughput, high-resolution approach is far more comprehensive and less biased than conventional techniques.
Customer Review
Unprecedented Site Coverage
"Using Creative Biolabs' ubiquitination modification proteomic quantitative analysis service in our research has significantly improved our ability to map ubiquitylation sites. Their use of complementary antibodies and high-resolution MS platforms provided an unprecedented depth of coverage, revealing novel ubiquitylation events on previously uncharacterized proteins. The data quality was exceptional, far exceeding our in-house capabilities." - Dr. A. Wri***t.
Reliable Quantitative Insights
"We needed to precisely quantify ubiquitylation changes in response to a novel proteasome inhibitor. Creative Biolabs's service delivered highly reproducible quantitative data using TMT labeling, allowing us to confidently identify dose-dependent ubiquitylation changes. Their detailed statistical analysis and clear data presentation were invaluable for our drug development pipeline." - Prof. D. Gre***n.
Reference
-
Wagner, Sebastian A., et al. "Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues." Molecular & Cellular Proteomics 11.12 (2012): 1578-1585. DOI: 10.1074/mcp.M112.017905. Distributed under an Open Access license CC BY 4.0, without modification.
Related Services
For Research Use Only.
 Contact Us
Follow us on
Contact Us
Follow us on
 Fig.1 Biological processes associated with ubiquitinated proteins.1
Fig.1 Biological processes associated with ubiquitinated proteins.1

 Fig.2 A protocol for the analysis of endogenous ubiquitination sites in mouse tissues using MS.1
Fig.2 A protocol for the analysis of endogenous ubiquitination sites in mouse tissues using MS.1