Lipid-based drug delivery systems, known for their ability to encapsulate both hydrophobic and hydrophilic molecules, are extensively used to improve the bioavailability of poorly soluble drugs and to protect them from degradation in vitro and in vivo. The average particle size and polydispersity index (PDI) are critical determinants of their safety, stability, efficacy, and behavior in biological environments.
The suitability of lipid-based drug delivery systems for specific routes of administration depends on parameters like average particle size/diameter and PDI. These factors also affect the processability, appearance, volume, and stability of the final pharmaceutical product. Therefore, controlling and validating these parameters is crucial for their effective clinical use.
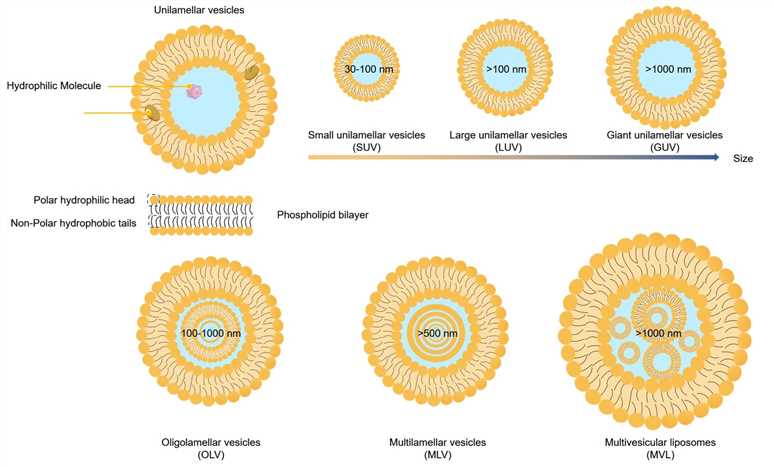 Fig.1 Classification of lipid-based drug delivery systems according to their particle size and lamellarity.
Fig.1 Classification of lipid-based drug delivery systems according to their particle size and lamellarity.
Particle size is a pivotal parameter in lipid-based drug delivery systems, affecting drug release, encapsulation efficiency, stability, biodistribution, adhesion, and the uptake by cells . Smaller particles generally have a higher surface-to-volume ratio, which can enhance solubility, increase bioavailability, improve controlled release, and allow precise drug targeting.
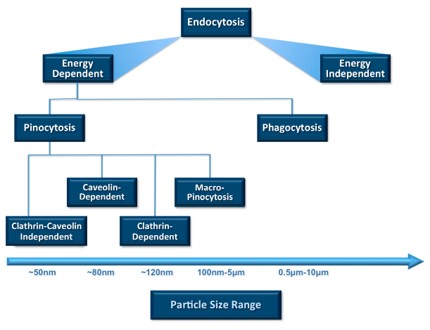 Fig.2 Particle sizes favorable for cellular uptake and ingestion through endocytotic pathways.1
Fig.2 Particle sizes favorable for cellular uptake and ingestion through endocytotic pathways.1
The accumulation of nanoparticles, such as liposomes, in target tissues is influenced by their particle size distribution, with a uniform distribution being conducive to the preparation of stable, safe, and efficient liposomes. The PDI quantifies nanoparticle size distribution, expressing size variability on a scale from 0 to 1. In the context of liposomal drug delivery, a PDI of 0.3 or lower is considered acceptable, indicating a homogeneous population of liposomes, whereas a higher PDI suggests a heterogeneous sample with a broad size distribution.
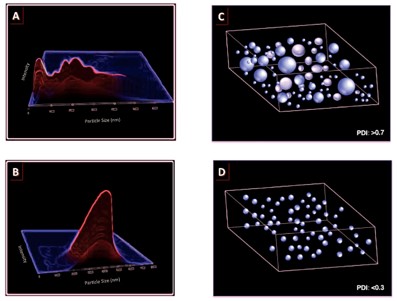 Fig.3 The relationship between particle size distribution and PDI value.1
Fig.3 The relationship between particle size distribution and PDI value.1
Successfully prepared liposomes can be downsized using additional techniques such as sonication, high-pressure homogenization, or extrusion.
Sonication methods, including bath and probe sonication, deliver high-energy inputs based on cavitation to liposome solutions within a passive (inert) atmosphere. While effective, this method can cause vesicle rupture, and probe sonication might release metal particles, risking contamination.
Liposomes can be downsized via high-pressure homogenization, which forces them through an orifice under high pressure. The resulting size distribution is influenced by variables such as temperature, pressure, the number of homogenization cycles, initial liposome size distribution, ionic strength, lipid composition, and lamellarity, which can lead to a broad and variable range.
Extrusion is performed post-liposome formation, where liposomes are passed repeatedly through a membrane with a specific pore size, often using polycarbonate filters, to achieve a uniform size distribution. This method does not require high pressure, facilitating reproducible outcomes for the final liposome product, but it has scalability limitations for large-scale manufacturing.
Various techniques can be employed to assess the average size and size distribution of liposomes, including microscopy, diffraction and scattering techniques, and hydrodynamic methods.
Microscopy, which includes optical, scanning electron (SEM), and transmission electron microscopy (TEM), plays a crucial role in assessing shapes, particle size, structures, surface properties, and stability. However, these methods provide only semi-quantitative analysis of particle size distribution from images and may fail to detect large aggregates or ultra-small particles when their concentration is low. Additionally, these techniques often require complex sample preparation, which is time-consuming and not suitable for routine particle size analysis.
DLS, also known as photon correlation spectroscopy (PCS), is esteemed for its rapid, straightforward, and reliable method for evaluating liposome size and PDI. It measures the intensity of light scattered by particles undergoing Brownian motion as a function of time. This technique is valued for its ease of operation, minimal sample requirement, reproducibility, and full automation, making it the mainstream approach for convenient and quantitative analysis of nanoparticle size and PDI within the 0.5-1000 nm diameter range.
Several additional analytical techniques enable the assessment of nanoparticle size and PDI. Scanning ion occlusion sensing (SIOS) is capable of analyzing nanoparticles from 60 nm to several micrometers in size. Flow cytometry (FCM) is applicable for analyzing MLV or LUV, necessitating the fluorescent labeling of samples to distinguish them from impurities and noise signals. Nanoparticle tracking analysis (NTA) allows for the tracking and measurement of individual nanoparticles under Brownian motion, effectively measuring particle size, size distribution, and concentration within the 30-1000 nm range.
Creative Biolabs specializes in precise liposome particle size measurement, offering advanced characterization services to ensure your formulations meet the highest standards. Contact us today for expert support and optimization of your liposome performance. Partner with us for excellence in liposome science.
Reference
 For Research Use Only. Not For Clinical Use
For Research Use Only. Not For Clinical UseSupports
Online Inquiry