N-Linked Glycoengineering Service in Yeast
N-Glycosylation in Yeast and Mammals
Yeast and other mammalian cells share similar initial biosynthetic pathways for crafting N-glycans. Briefly, Glc3Man9GlcNAc2, a lipid-linked oligosaccharide (LLO), is synthesized within the endoplasmic reticulum (ER) through the concerted action of various glycosyltransferases. This LLO is subsequently attached to asparagine residues on the emerging polypeptide chain and trimmed down to Man8GlcNAc2. The newly formed glycoconjugate is then transported to the Golgi apparatus. Further glycan processing in the Golgi differs between yeast and mammalian cells. Mammalian cells employ α-mannosidases to further trim the Man8GlcNAc2 N-glycan, yielding a substrate that allows for the creation of hybrid- and complex-type N-glycans via glycosyltransferases. In contrast, yeast cells elongate the existing high mannose structure by adding extra mannose sugars, ultimately resulting in the production of hyper-mannosylated glycans.
N-Linked Glycoengineering Services in Yeast at Creative Biolabs
With a wealth of experience in Cell Line Glycoengineering Services, Creative Biolabs has implemented multiple glycoengineering strategies and genetic techniques, such as Knockdown, Knockout, and Transient Overexpression, to enhance the yeast N-glycosylation pathway for the synthesis of recombinant glycoproteins and therapeutic proteins with uniform and humanized glycans. After eliminating undesirable yeast-specific glycan structures, we focus on humanizing the N-linked glycosylation pathway in yeast. This includes introducing the necessary glycosidases and glycosyltransferases to enable the production of hybrid- and complex-type glycans, closely resembling human-type glycosylation patterns.
Removal of yeast-specific N-glycans
-
Elimination of yeast glycosyltransferase such as Och1p, MNN1.
-
Knockout of Bmt2p for β-Mannose depletion.
-
Intervention in the LLO assembly.
-
Overexpression of Endo-β-N-Acetylglucosaminidases (ENGases).
Humanization of the N-glycosylation pathway
-
Overexpression of human N-acetylglucosaminyltransferases, such as GnT-I, GnT-II.
-
Overexpression of β-1,4-galactosyltransferase (GalT) in Golgi.
-
Introduction of sialyltransferase (ST) for terminal sialylation.
-
Improvement of N-glycan site occupancy.
Features of Our Services
-
Multi-targeted and optional glycoengineering strategies
-
Highly efficient and precise knockin or knockout of genes
-
High-quality glycoprotein production with fully humanized N-glycans
-
Customized glycoengineering tailored to specific needs
Published data
This study focused on the potential of diploid Pichia pastoris in the expression of foreign proteins and its stability in high-yield fermentation. Using anti-HER2 monoclonal antibodies as targets, the authors demonstrated that both wild-type and glycoengineered diploid strains could effectively and stably produce recombinant proteins through nutrient-rich shake flask cultures. Although spore formation was observed in the bioreactor, it did not significantly affect production efficiency, and the recombinant antibodies produced by the diploid strains were of good quality, with N-glycosylation profiles and protein production capacity comparable to those of haploid strains. This study systematically demonstrated for the first time the potential of diploid P. pastoris for the expression and fermentation of recombinant proteins, supporting further development and optimization of mating technology in areas such as strain integration and antibody display.
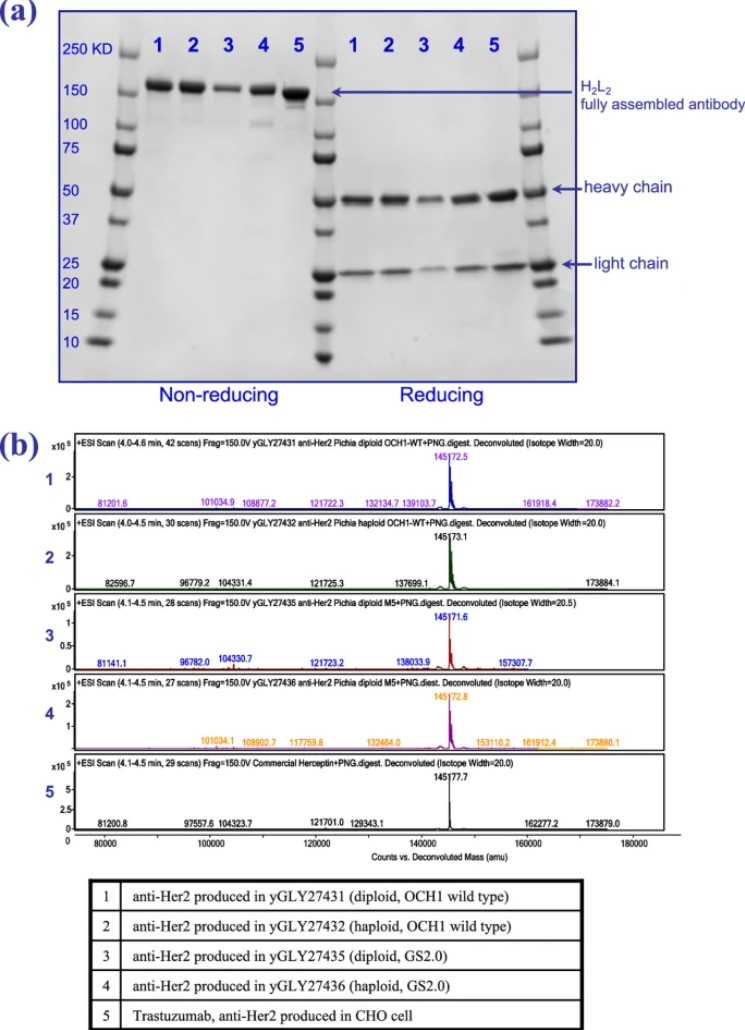 Fig.1 Characterization results of diploid P. pastoris-derived antibodies.1
Fig.1 Characterization results of diploid P. pastoris-derived antibodies.1
Scientists at Creative Biolabs have amassed a wealth of expertise in the field of glycoengineering in yeast. We are committed to offering a full spectrum of customized glycoengineering services, tailored to meet your unique needs. If you have any special requests related to glycoengineering, we encourage you to contact us without hesitation.
Reference
-
Chen, Ming-Tang, et al. "Generation of diploid Pichia pastoris strains by mating and their application for recombinant protein production." Microbial Cell Factories 11 (2012): 1-18. Distributed under Open Access license CC BY 2.0, without modification.
For Research Use Only.
Related Services
 Fig.1 Characterization results of diploid P. pastoris-derived antibodies.1
Fig.1 Characterization results of diploid P. pastoris-derived antibodies.1