Structure Prediction of Fv & CDR
It is of great significance to predict the structure of Fv and CDRs, the core structure of an antibody, for novel antibody design and engineering. Based on various structure-prediction tools such as RosettaAntibody, PIGS, and SAbPred, Creative Biolabs is able to provide a full range of solutions for structure prediction of Fv and CDRs to accelerate the development and engineering of novel antibody.
Introduction of Structure Prediction of Fv and CDRs
It is very important to predict the three-dimensional structure of an antibody for antibody engineering including optimizing antibody affinity, specificity, and stability. The most significant part of antibody structure prediction is to predict the structure of antibody variable fragment (Fv) because the constant regions of an antibody are virtually similar in all antibodies of the same isotype. Fv is composed of two chains, heavy variable domain (VL) and light variable domain (VH), and each of which contains three hypervariable complementarity-determining regions (CDRs) that determine the interactions with antigens. Because of the extremely conserved nature of the framework regions of the VL and VH domains, the research on Fv structure prediction is mainly focused on the prediction of 6 CDR loops.
Currently, many antibody structure-prediction methods have been developed, which use sequence homology to natural antibody structures to select backbones for modeling. Besides, some automated web-servers, such as RosettaAntibody, PIGS, and SAbPred have also been established for structure-based predictions for antibody engineering and design. In general, the workflow of web-servers includes identifying the segments corresponding to the CDRs and framework in the query sequence, template structures from the Protein Data Bank (PDB) are then selected for each segment by sequence similarity and the resulting structure model is refined by energy minimization.
Several Automated Web-servers
-
SAbPred
SAbPred is a free web server that is used for the annotation of the sequences of antibodies with different numbering schemes, automatic generation and annotation of homology models of antibody Fv regions, prediction of antibody paratope residues, and prediction of antigen epitope residues. This tool has been used for structure-based predictions in antibody engineering and design. Importantly, SAbPred can automatically model the Fv structure of an antibody using its ABodyBuilder algorithm.
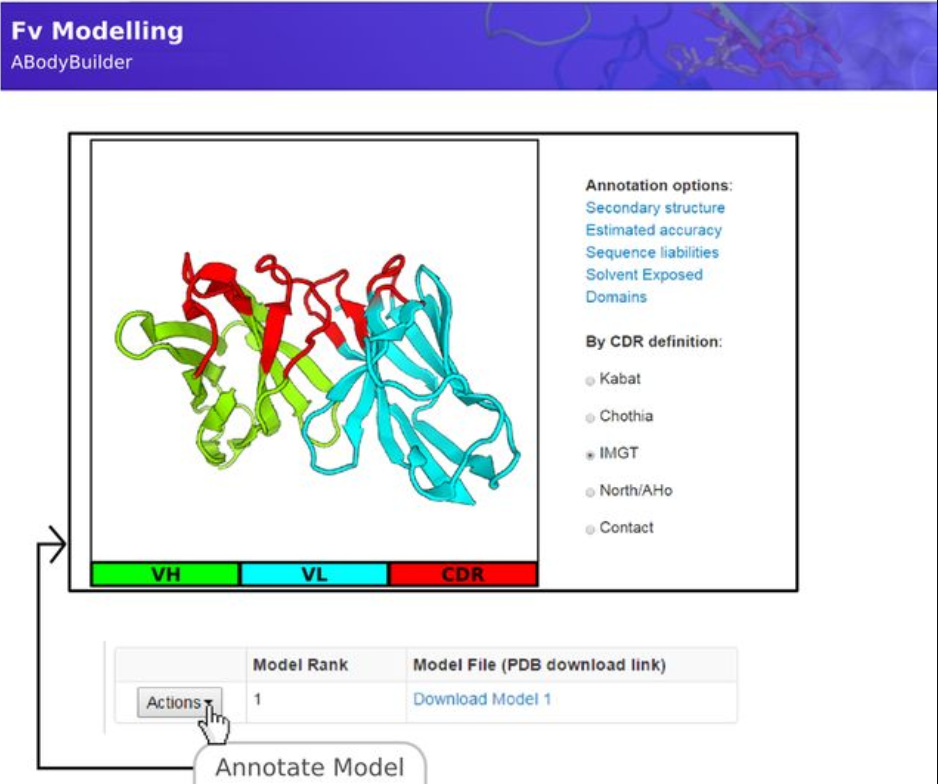 Fig. 1 The ABodyBuilder tool is an automatic Fv modeling protocol.1
Fig. 1 The ABodyBuilder tool is an automatic Fv modeling protocol.1
-
PIGS (Prediction of Immunoglobulin Structure)
PIGS server is a flexible and user-friendly tool to model the structure of immunoglobulins. It relies on a database of known immunoglobulin structures and of their structural alignment. The sequence of the variable chains of the antibody of interest is input on the server and the program will output a list of putative templates for both the loops and the framework for each chain. The user can select the templates for the frameworks and the loops using different strategies. Finally, the program will output a complete 3D model of the target immunoglobulin variable domains.
-
RosettaAntibody
RosettaAntibody server allows prediction of the structure of an antibody variable region according to the given the amino-acid sequences of the respective light and heavy chains. In the first stage, the most sequence homologous template structures for the framework regions and each of the CDR loops can be identified by the server. Then, the template CDRs are grafted onto the framework and the side chains of all the residues in the assembled model are optimized, generating a side-chain optimized crude model. In the second stage, the server starts with a multi-start, multi-scale Monte Carlo-plus-minimization algorithm to produce two thousand candidate structures.
What Can We Do?
Creative Biolabs takes advantage of various antibody structure-prediction methods to help customers predict the structure of Fv and CDRs, promoting their antibody engineering programs. With extensive experience in the prediction of antibody structure, we have the capability of customizing different strategies for structural prediction of various antibodies according to the specific requirements from the customers. We ensure to deliver the optimal prediction structure models of Fv and CDRs models which can be used for designing and engineering antibodies with desired properties.
If you are interested in antibody engineering based on the structure prediction of Fv and CDRs, please feel free to contact us for more details.
Reference
- James, D.; et al. SAbPred: a structure-based antibody prediction server. Nucleic Acids Research. 2016, 44(W1): W474-W478. Distributed under Open Access license CC BY 4.0, without modification.
For Research Use Only.
