N-glycosylation represents an essential type of post-translational modification (PTM) involving the attachment of oligosaccharides to asparagine (Asn) residues located in the Asn-X-Ser/Thr motif (X ≠ Pro). The endoplasmic reticulum and Golgi apparatus host this process which deeply influences protein folding stability trafficking and functionality. Researchers can access our customized suite of N-glycosylation analysis services at Creative Biolabs. We have a dedicated staff of seasoned scientists who possess extensive knowledge of glycosylation biology and analytical methods and they are dedicated to delivering outstanding service. The experts possess comprehensive knowledge about recent research progress in their field and deliver significant support throughout every stage of analysis including sample preparation and data interpretation. We have developed our services to carefully analyze the intricate aspects of N-glycosylation!
Why N-Glycosylation Matters
N-glycosylation regulates essential biological processes:
-
Protein folding and quality control
-
Cell-cell interactions and immune responses
-
Disease mechanisms like cancer metastasis
Understanding these dynamics requires cutting-edge analytical strategies—something we specialize in. Our services combine advanced MS platforms, customizable workflows, and deep expertise to decode glycosylation patterns efficiently.
Four Levels of N-Glycosylation Analysis
This approach examines intact glycoproteins to capture their full glycoform profiles. High-resolution mass spectrometry (HRAM-MS) reveals mass variations and glycan compositions. While it provides a broad overview, resolving site-specific details remains challenging. Our HRAM-MS platforms deliver unparalleled resolution, ideal for characterizing complex biotherapeutics like monoclonal antibodies (mAbs).
Glycopeptide Analysis
By enzymatically digesting glycoproteins, we pinpoint glycosylation sites and assess heterogeneity. LC-MS/MS with HCD or EThcD fragmentation maps site-specific glycan structures. Customized enzymatic workflows and AI-driven data tools streamline glycopeptide identification, even in dense biological matrices.
Released Glycan Analysis
PNGase F releases N-glycans for structural profiling. Techniques like HILIC-FLD or MALDI-TOF/MS separate and quantify glycans, though site linkage data is lost. We offer rapid derivatization (e.g., 2-AB labeling) and PGC-LC-MS workflows to resolve isomers, critical for biomarker studies.
Acid hydrolysis breaks glycans into monosaccharides, analyzed via HPAE-PAD or GC-MS. This reveals sugar types/quantities but not linkages. Pair this with glycan linkage analysis for a complete picture—our GC-MS platform ensures precision down to trace levels.
N-Glycosylation Analysis Workflow
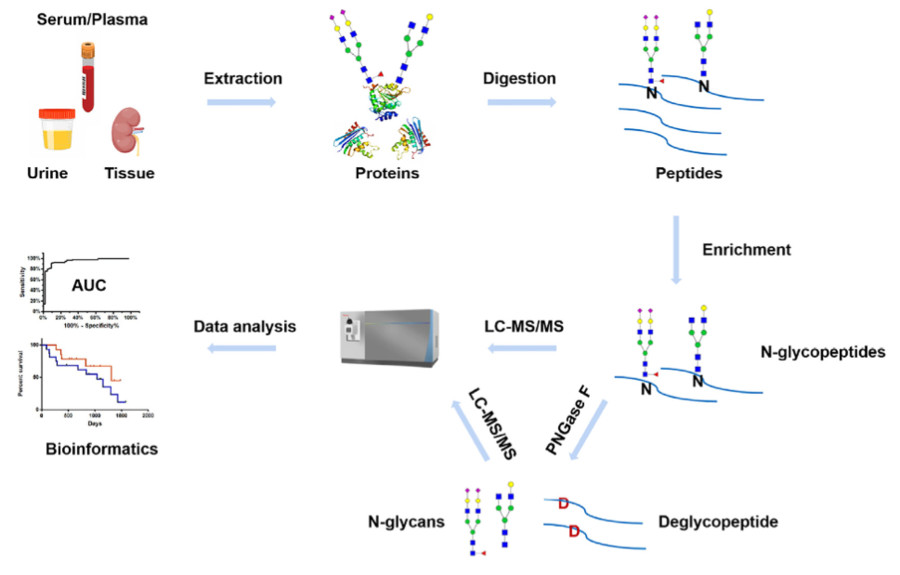 Fig.1 Workflow of MS-based N-glycosylation analysis in clinical samples.1
Fig.1 Workflow of MS-based N-glycosylation analysis in clinical samples.1
1. Sample Preparation
-
Biotherapeutics: CHO cell cultures, mAb purification.
-
Clinical Samples: Serum/urine/tissue samples protein extraction, impurity removal (SPE, ultrafiltration).
If you need help with tricky samples, please make a request and our team are committed to optimizing protocols for low-abundance targets.
2. Glycan Release
-
Enzymatic (PNGase F) or chemical cleavage.
-
Denaturation steps (SDS) boost efficiency.
3. Labeling & Derivatization
-
Fluorescent tags (2-AB) for HILIC-FLD.
-
Useful chemical reagents for MS-friendly rapid processing.
4. Separation
-
HILIC, RP-HPLC, or CE—we match techniques to your throughput needs.
5. MS Analysis
-
MALDI-TOF MS: Quick glycan profiling.
-
LC-MS/MS: Site-specific mapping via Orbitrap Fusion Tribrid MS.
-
High-Res MS: Resolve microheterogeneity with sub-ppm accuracy.
6. Data Interpretation and Reporting
-
Utilize tools to decode spectra and structural information.
-
Ensure consistent results through method validation and inter-laboratory comparisons.
Site-Specific Analysis & Prediction
Human N-glycosylation hinges on the Asn-X-Ser/Thr motif, but structural context matters. If you are studying a therapeutic protein, our site-specific analysis ensures batch consistency and minimizes immunogenicity risks. Our workflows integrate:
-
Labeling: Stable isotope labeling with amino acids in cell culture (SILAC), tandem mass tag (TMT) and isobaric tags for relative and absolute quantitation (iTRAQ) for quantitative comparisons across conditions.
-
Analytical Techniques
-
Mass Spectrometry (MS): High-resolution MS enables the identification and quantification of glycopeptides and glycan structures.
-
Liquid Chromatography (LC-MS/MS): Combines separation capabilities with MS for detailed glycan profiling.
-
Enzymatic Digestion: Enzymes like PNGase F are used to release N-linked glycans from glycoproteins for analysis.
-
Prediction Tools: Tools are trained on human proteome data.
Real-World Applications
-
Biopharma Development
Glycosylation affects drug stability and efficacy. Our services ensure your mAbs meet regulatory standards—fast.
-
Disease Mechanisms
Uncover glycosylation shifts in cancer or neurodegeneration. We've helped clients identify novel biomarkers in <6 weeks.
-
Diagnostics
Aberrant serum glycans signal disease. Partner with us to develop LC-MS assays for early detection.
Why Partner with Creative Biolabs?
-
End-to-End Solutions: From sample prep to bioinformatics—no outsourcing hassles.
-
Cutting-Edge Tech: Orbitrap Fusion Tribrid MS, CE, and custom HILIC columns.
-
Speed & Precision: High-throughput workflows with 98% reproducibility.
Our N-Glycosylation Analysis Services provide actionable data for biotherapeutic profiling and disease biomarker exploration. Reach out to us for a no-cost consultation to learn how we can speed up your project.
FAQs
1. How to analyze glycosylation?
Analyzing glycosylation requires multiple sequential stages. First, glycans are released from glycoproteins. The release process can utilize enzymes such as PNGase F or proceed through chemical methods. Next is the labeling step. For labeling purposes mass spectrometry compatible fluorescent dyes or reagents are frequently utilized. Then, separation techniques come into play. Researchers commonly use hydrophilic interaction liquid chromatography (HILIC), reversed-phase high-performance liquid chromatography (RP-HPLC), or capillary electrophoresis as separation methods during glycosylation analysis. The final analytical step involves using mass spectrometry to identify and quantify the analytes. Through this complete approach researchers achieve an intricate understanding of glycan structures along with their biological functions.
2. What is the N-glycosylation in diabetes?
In type 2 diabetes patients, researchers can observe distinct alterations in protein glycosylation patterns. Altered protein function from these changes accelerates the disease development process. Changes in the N-glycosylation patterns of immunoglobulin G (IgG) correlate with a higher likelihood of developing type 2 diabetes. Glycosylation patterns may develop into biomarkers which predict disease susceptibility.
3. What role does glycosylation play in defining human blood type?
Specific monosaccharides present or absent on red blood cell surfaces determine blood type. The attachment of N-acetylgalactosamine to the O antigen forms type A blood while the addition of galactose generates type B blood. These modifications are catalyzed by specific glycosyltransferases. The distinct ABO blood types are produced through variations in enzyme activity.
Reference
-
Ren, Weifu, Qi Bian, and Yan Cai. "Mass spectrometry-based N-glycosylation analysis in kidney disease." Frontiers in Molecular Biosciences 9 (2022): 976298. Distributed under Open Access license CC BY 4.0, without modification. https://doi.org/10.3389/fmolb.2022.976298
Resources
For Research Use Only.
 Contact Us
Follow us on
Contact Us
Follow us on
 Fig.1 Workflow of MS-based N-glycosylation analysis in clinical samples.1
Fig.1 Workflow of MS-based N-glycosylation analysis in clinical samples.1