Are you currently facing challenges in identifying novel, highly specific biomarkers for ovarian cancer, or seeking to understand the intricate mechanisms of cancer progression at a molecular level? Creative Biolabs' Glycosylation Analysis Services for diseases help you accelerate biomarker discovery and deepen mechanistic insights through advanced glycomics and glycoproteomics technologies.
How Creative Biolabs' Ovarian Cancer related Glycosylation Analysis Service Can Assist Your Project?
Creative Biolabs stands at the forefront of glycosylation analysis, driven by over two decades of specialized expertise in advanced glycomics and glycoproteomics. We provide specialized solutions to unravel the complex role of glycosylation in ovarian cancer. We deliver precise, quantifiable data on aberrant glycan structures and modified glycoproteins, enabling the identification of novel diagnostic and prognostic biomarkers, as well as potential therapeutic targets. Our service helps you move beyond traditional protein-level analysis to discover site-specific glycosylation changes, which are often more sensitive and specific indicators of disease. We assist in elucidating the functional impact of these glycan alterations on key cellular processes such as cell signaling and protein transport, offering a deeper understanding of ovarian cancer biology.
Key Steps in Ovarian Cancer related Glycosylation Analysis Services
-
Activity: Extraction and purification of proteins, typically membrane glycoproteins or total serum proteins, from provided biological samples. This step is crucial for isolating the target proteins for subsequent glycan analysis, ensuring high purity and integrity.
-
Outcome: High-quality protein extracts ready for targeted glycan release.
-
Activity: N-linked glycans are enzymatically released (e.g., using PNGase F), and O-linked glycans are chemically released (e.g., via β-elimination). Sialic acid residues are chemically stabilized to prevent loss during mass spectrometry. Reduced ends of glycans may be labeled with isobaric tags for quantitative analysis across multiple samples.
-
Outcome: Purified and derivatized glycan, which is suitable for high-resolution mass spectrometry.
-
Activity: Application of advanced mass spectrometry platforms (e.g., LC-ESI-MS, MALDI-TOF MS) for comprehensive glycan and glycopeptide profiling. This involves fragmenting glycans to determine their precise structural details and quantifying their relative abundance.
-
Outcome: Raw mass spectrometry data containing detailed information on glycan compositions, structures, and relative quantification.
-
Activity: Sophisticated bioinformatics tools and databases are used to interpret MS/MS spectra, assign glycan structures, and accurately quantify changes in specific glycan populations between cancer and control samples.
-
Outcome: Identified and quantified N-linked and O-linked glycan structures, highlighting those with significant differential expression.
-
Activity: Identified glycoproteins are analyzed using bioinformatics tools to annotate their known functions and involvement in biological pathways relevant to ovarian cancer (e.g., Notch, Wnt).
-
Outcome: Insights into the potential functional roles of aberrantly glycosylated proteins in ovarian cancer progression.
-
Comprehensive glycan profiling reports: Detailed lists and relative quantification of all identified N-linked and O-linked glycan structures, highlighting statistically significant differences.
-
Glycoprotein/Glycopeptide identification reports: For glycoproteomics projects, comprehensive lists of identified glycoproteins and their associated glycosylation sites.
-
Bioinformatics analysis and functional insights: Interpretation of findings, including correlations between glycan changes, gene expression, and potential implications for disease mechanisms.
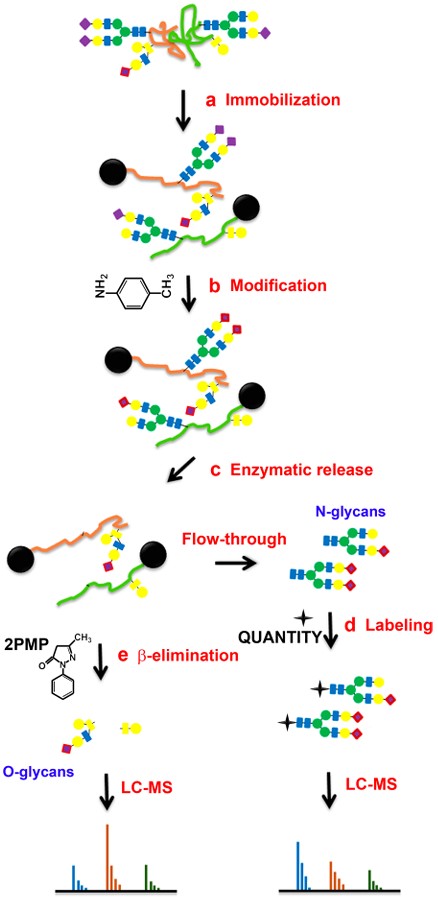 Fig.1 Release and analysis of N-linked and O-linked glycans.1
Fig.1 Release and analysis of N-linked and O-linked glycans.1
What Is the Importance of Glycosylation Analysis in Ovarian Cancer Research?
Glycosylation analysis is paramount in ovarian cancer research due to the disease's high mortality rate and the limitations of current diagnostic markers like CA125. Cancer cells often exhibit altered glycosylation patterns, which are not merely consequences of malignancy but actively contribute to tumor progression, invasion, and immune escape. By precisely characterizing these changes, we can:
-
Improve early detection
-
Uncover disease mechanisms
-
Develop targeted therapies
-
Monitor disease progression and treatment response
In conclusion, Creative Biolabs' ovarian cancer related glycosylation analysis service offers a transformative approach to understanding and combating ovarian cancer. Leveraging over 20 years of expertise and state-of-the-art glycomics and glycoproteomics technologies, we provide unparalleled precision in identifying novel, site-specific glyco-biomarkers and elucidating the complex functional roles of aberrant glycosylation. Our comprehensive services, from detailed glycan profiling to insightful biological interpretation, empower researchers and biopharmaceutical companies to accelerate biomarker discovery, deepen mechanistic understanding, and pave the way for next-generation diagnostics and therapeutics. Contact our team today for more information and to discuss your project. Our experts are eager to provide a tailored solution that meets your specific research needs and accelerates your breakthroughs.
Published Data
This research paper introduces an innovative solid-phase chemoenzymatic methodology designed for the concurrent analysis of N-linked and O-linked glycans found in ovarian cancer cells. The method involves immobilizing cellular proteins, chemically protecting sialic acid residues to prevent their degradation, followed by the sequential release of N-glycans using an enzyme and O-glycans via a chemical process. A significant enhancement of the approach is the improved detection of fragile sialylated glycans due to their stabilization. The study applied this platform to investigate the impact of an O-linked glycosylation inhibitor, Benzyl-α-GalNAc (BAG), on the glycan profiles of OVCAR-3 ovarian cancer cells. The results showed that lower concentrations of the O-linked glycosylation inhibitor appeared to increase the levels of N-glycans. This suggests a complex interplay and potential regulatory link between the biosynthesis pathways of these two major glycan types. Figure 2 serves as a crucial visual representation of the analytical power of their method regarding N-glycans.
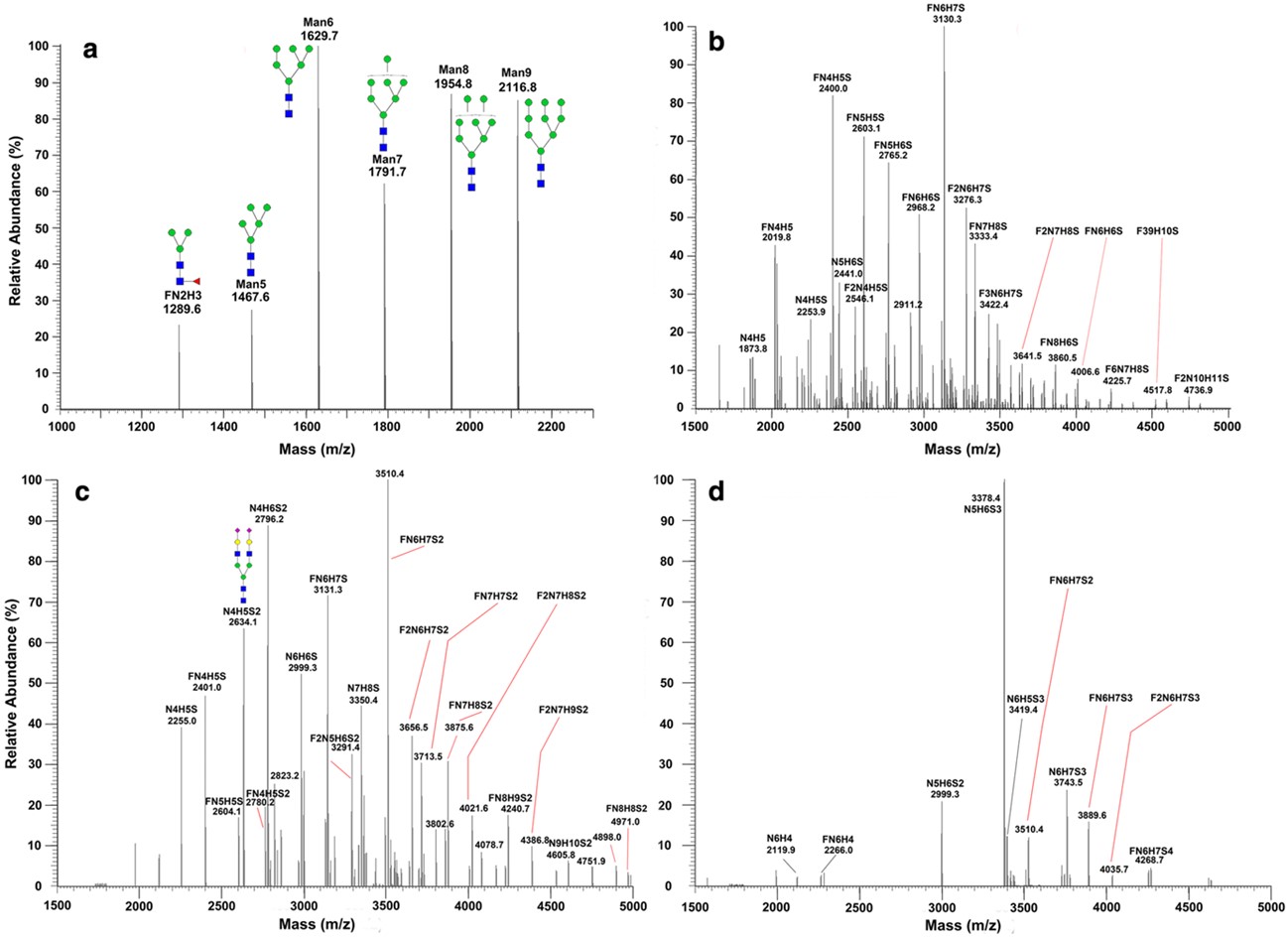 Fig.2 LC-ESI-MS analysis of OVCAR-3 ovarian cancer cells.1
Fig.2 LC-ESI-MS analysis of OVCAR-3 ovarian cancer cells.1
FAQs
Q1: What types of samples can Creative Biolabs analyze for ovarian cancer glycosylation?
A1: We are equipped to analyze a wide range of biological samples, including human serum, plasma, fresh/frozen ovarian tumor tissues, normal ovarian tissues, and various ovarian cancer cell lines. Our flexible sample preparation methods ensure optimal glycan recovery from each matrix. If you have specific sample types not listed, please reach out to our team for a personalized consultation!
Q2: How does Creative Biolabs ensure the accuracy and reliability of its glycosylation data?
A2: Our commitment to accuracy is paramount. We employ a multi-pronged approach that includes high-resolution mass spectrometry, stable isotope labeling for quantitative precision, and rigorous bioinformatics analysis with multiple validation steps. Our team of experienced glycomic scientists manually verifies critical glycan structures, and we utilize established quality control measures throughout the workflow to ensure highly reliable and reproducible results.
Q3: Beyond identifying glycan changes, can Creative Biolabs help us understand the biological significance or functional implications in ovarian cancer?
A3: Yes, our service extends beyond raw data. We integrate glycomic findings with bioinformatics analyses to map identified glycoproteins to relevant biological pathways (e.g., cell signaling, protein transport, immune modulation). We also offer insights into the correlation between glycan alterations and gene expression of glycosyltransferases, including epigenetic influences like DNA methylation, providing a deeper mechanistic understanding of how these changes contribute to ovarian cancer biology.
Customer Review
Comprehensive Data and Clear Interpretation
"Using Creative Biolabs' ovarian cancer research related glycosylation analysis service in our research has significantly improved our ability to distinguish between benign and malignant ovarian tumors based on specific N-glycan patterns, offering much-needed specificity compared to traditional markers. Their attention to detail in the data and clear interpretation were invaluable." - Dr. V. Mit***l.
Specialized Glycomic Analysis
"Creative Biolabs's glycoproteomics capabilities allowed us to pinpoint novel site-specific sialylation changes on serum proteins directly linked to ovarian cancer progression. This level of resolution, achieved efficiently through their advanced mass spectrometry, has opened up entirely new avenues for our therapeutic development program. Their team's expertise in interpreting the complex glycan structures was a major advantage." - Prof. X. Ric***s.
Reference
-
Yang, Shuang, et al. "Simultaneous analyses of N-linked and O-linked glycans of ovarian cancer cells using solid-phase chemoenzymatic method." Clinical Proteomics 14 (2017): 1-11. DOI: 10.1186/s12014-017-9137-1. Distributed under an Open Access license CC BY 4.0, without modification.
Related Services
For Research Use Only.
 Contact Us
Follow us on
Contact Us
Follow us on
 Fig.1 Release and analysis of N-linked and O-linked glycans.1
Fig.1 Release and analysis of N-linked and O-linked glycans.1
 Fig.2 LC-ESI-MS analysis of OVCAR-3 ovarian cancer cells.1
Fig.2 LC-ESI-MS analysis of OVCAR-3 ovarian cancer cells.1