Hepatitis C Virus (HCV) Envelope Glycans
Hepatitis C virus (HCV) remains a significant global health burden, affecting millions of individuals worldwide and leading to severe liver complications such as cirrhosis and hepatocellular carcinoma. A critical feature of HCV biology is its highly glycosylated envelope, which plays a dual role in facilitating viral entry and protecting the virus from the host immune system. The envelope glycoproteins, E1 and E2, are heavily coated with N-linked glycans that form a dense "glycan shield," masking conserved epitopes from neutralizing antibodies. Understanding the intricate architecture of these glycans is essential for designing effective immunogens and therapeutic agents. At Creative Biolabs, we offer specialized research services, including Anti-HCV Glycan Shield Antibody Development, to assist scientists in decoding the complex interplay between HCV envelope glycosylation and immune recognition.
Introduction to HCV Envelope Glycans
The HCV particle is composed of a nucleocapsid surrounded by a lipid bilayer derived from the host endoplasmic reticulum (ER), into which the viral envelope glycoproteins E1 and E2 are anchored. These glycoproteins are vital for the viral life cycle, mediating receptor binding and membrane fusion. A defining characteristic of E1 and E2 is their extensive post-translational modification by N-linked glycans. These carbohydrates account for nearly half of the molecular mass of the E1E2 heterodimer. The high density of glycans on the viral surface creates a dynamic barrier that modulates protein folding, stability, and viral entry while simultaneously serving as a primary mechanism for neutralizing antibody evasion.
Architecture of Hepatitis C Virus E1 E2 Glycoproteins
E1 and E2 interact to form a non-covalent heterodimer, which is believed to be the functional unit on the virion surface. Both proteins are type I transmembrane proteins with C-terminal transmembrane domains that facilitate membrane anchoring and heterodimerization.
E2 Glycoprotein
E2 is the receptor-binding subunit and the primary target for neutralizing antibodies. It contains approximately 11 N-linked glycosylation sites (depending on the genotype), many of which are highly conserved. These glycans are critical for the proper folding of the E2 protein and its subsequent interaction with E1. Structural studies have revealed that E2 possesses a central immunoglobulin-like β-sandwich core flanked by variable regions (HVR1, HVR2, and igVR), which are often shielded by the glycan canopy.
E1 Glycoprotein
E1 is smaller than E2 and contains 4 to 5 N-linked glycosylation sites. Although less well-characterized than E2, E1 is essential for membrane fusion. The glycans on E1 also contribute to the overall stability of the heterodimer and assist in masking the fusion peptide until the virus engages with the host cell.
Functional Roles of HCV N-Glycans
The dense array of N-linked glycans on HCV E1 and E2 is not merely a passive defense mechanism; it is intrinsic to the virus's structural integrity and infectivity. These glycans orchestrate critical steps in the viral life cycle, ranging from correct protein assembly in the endoplasmic reticulum to the fine-tuning of receptor interactions during entry. Their biological necessity makes them attractive targets for therapeutic intervention.
- Protein Folding and Heterodimerization: Specific glycans, particularly those at sites N196, N234, and N301 (H77 numbering), are indispensable for the recruitment of ER chaperones like calnexin and calreticulin. This interaction ensures the proper folding of the E1E2 heterodimer, preventing the formation of non-functional aggregates.
- Modulation of Viral Entry: Glycans on E2, such as N417 and N532, act as "gatekeepers" that regulate the affinity of the virus for the CD81 receptor. By masking the CD81 binding site, these glycans prevent premature activation and ensure the virus only engages its receptor when in the correct cellular context.
- Lectin-Mediated Capture: The abundance of high-mannose glycans on the viral envelope facilitates interaction with C-type lectins (e.g., DC-SIGN and L-SIGN) expressed on dendritic cells and liver sinusoidal endothelial cells, promoting viral capture and transmission to hepatocytes.
The Glycan Shield as a Therapeutic Target
While the primary function of the glycan shield is to evade immune surveillance, its conservation across diverse HCV genotypes creates a unique vulnerability. Unlike the hypervariable peptide regions, the positioning of many N-glycans is highly conserved, presenting a stable target for therapeutic design.
Glycan-Dependent Neutralizing Antibodies
Recent studies have isolated potent broadly neutralizing antibodies (bNAbs) that do not just bind to the protein surface but interact directly with the glycan shield. These antibodies target "glycan-dependent epitopes," where the glycan forms an integral part of the antibody binding site. Targeting these complex epitopes offers a strategy to overcome viral escape mutants that rely on glycan shifting.
Rational Immunogen Design
Understanding the shield's topology allows for "glycan engineering" of vaccine candidates. By selectively removing specific glycans (e.g., N276), researchers can expose conserved neutralizing epitopes (like the CD81 binding site) to the immune system, thereby eliciting a more potent neutralizing antibody response. Conversely, retaining key glycans ensures the immunogen mimics the native viral structure.
Our Solutions for HCV Glycan Research
Creative Biolabs provides a comprehensive suite of services to support the characterization of viral envelope glycans and the development of novel therapeutics. Our platform integrates advanced mass spectrometry, structural biology, and immunological assays to dissect the HCV glycan shield.
Anti-HCV Glycan Shield Antibody Development
We specialize in generating antibodies that target the HCV glycan shield or glycan-dependent epitopes. Utilizing our proprietary phage display libraries and hybridoma platforms, we can isolate bNAbs capable of penetrating the glycan dense layer or recognizing specific glycoforms essential for viral fitness.
Glycoarray Screening
Our glycoarray platforms enable high-throughput screening of viral envelope proteins against hundreds of defined glycan structures. This service is essential for identifying specific lectin interactions and characterizing the carbohydrate-binding profile of HCV E1E2.
Viral Glycosylation Analysis
We offer detailed mapping of N-linked and O-linked glycosylation sites on HCV envelope proteins using LC-MS/MS. Our analysis determines site occupancy, glycan composition (high-mannose vs. complex), and heterogeneity, providing a structural basis for HCV vaccine immunogen design.
Custom Glycosylation of Immunogens
To study the effect of specific glycans on immunogenicity, we provide custom expression services to produce E1E2 proteins with defined glycosylation patterns (e.g., oligomannose-only or complex-type enriched) using glyco-engineered cell lines.
Inquire About HCV Glycan Services
Published Data
A pivotal 2023 study highlights the functional consequences of "glycan shifting" on viral neutralization. The researchers evaluated the efficacy of antibodies raised against the conserved Epitope I (residues 412–425) using a cell culture-derived HCV (HCVcc) system. The neutralization data revealed a critical immune evasion mechanism: while antibodies elicited by the wild-type immunogen potently neutralized the wild-type virus (H77:WT), they completely failed to neutralize viral variants carrying the N417S mutation (H77:N417S). This mutation shifts a glycosylation site to position N415, structurally masking the neutralizing epitope. These findings, illustrated in the figure below, conclusively demonstrate that the shifting of a single glycan is sufficient to render potent antibodies ineffective, emphasizing the need for next-generation immunogens designed to overcome dynamic glycan shielding.
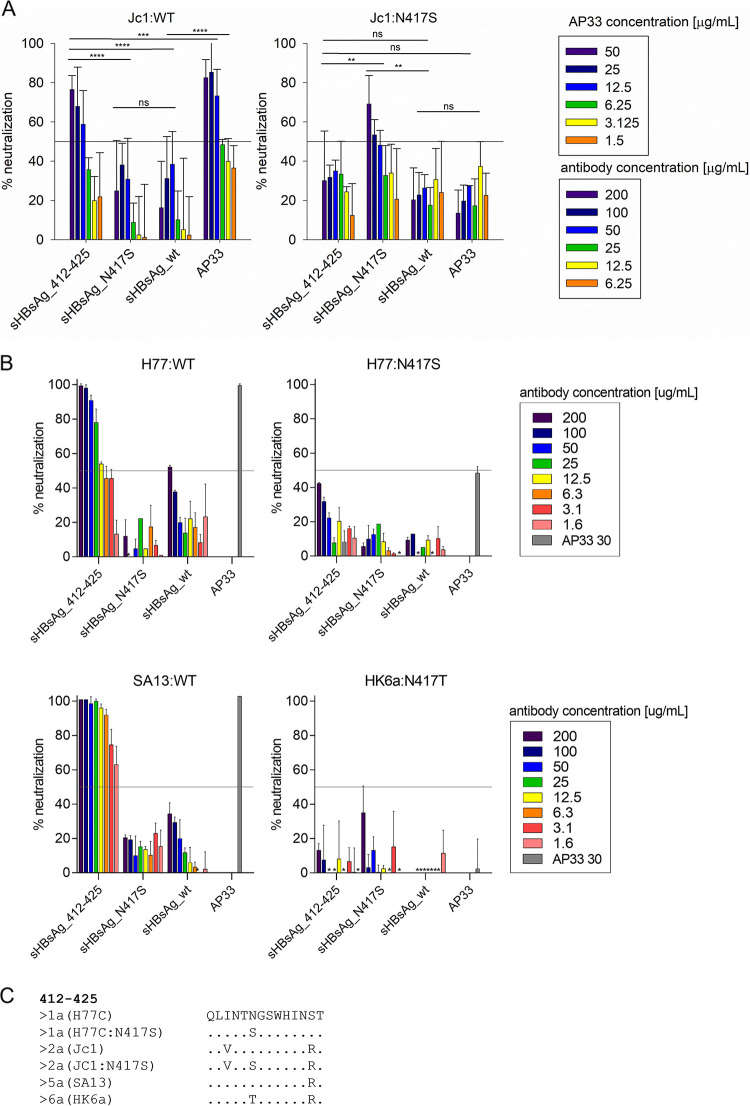 Fig.1
Impact of N417S Glycan Shift on HCV Neutralization.1
Fig.1
Impact of N417S Glycan Shift on HCV Neutralization.1
FAQs
Why is the glycan shield important for HCV vaccine development?
The glycan shield masks conserved epitopes that are necessary for viral neutralization. An effective vaccine must elicit antibodies that can either penetrate this shield or recognize the conserved regions that are transiently exposed during viral entry. Understanding the glycan shield layout helps in designing immunogens that present these vulnerable sites more effectively.
What is the difference between high-mannose and complex glycans on HCV?
High-mannose glycans are simpler structures often found in regions of the protein that are sterically restricted or difficult for processing enzymes to access. Complex glycans are fully processed in the Golgi. HCV E2 contains a mixture of both, and the specific composition can influence how the virus interacts with cellular lectins and antibodies.
How does Creative Biolabs analyze HCV envelope glycosylation?
We utilize a combination of proteolytic digestion and LC-MS/MS (Liquid Chromatography-Tandem Mass Spectrometry) to map glycosylation sites and determine the glycan structures present at each site (site-specific glycosylation profiling). We also use lectin microarrays for high-throughput screening of glycan-binding profiles.
Can you produce HCV pseudoparticles (HCVpp) with modified glycans?
Yes, we can generate HCVpp or cell culture-derived HCV (HCVcc) bearing specific mutations in N-linked glycosylation sites. This allows researchers to study the impact of individual glycans on viral infectivity, receptor binding, and neutralization sensitivity.
Do glycans affect the binding of the virus to CD81?
Yes, certain glycans on E2 act as a gatekeeper, modulating the interaction with CD81. Removing these glycans often increases the binding affinity to CD81, suggesting that they shield the receptor-binding site to prevent premature attachment or immune recognition.
Reference:
- Czarnota, A., et al. "Effect of Glycan Shift on Antibodies against Hepatitis C Virus E2 412–425 Epitope Elicited by Chimeric sHBsAg-Based Virus-Like Particles." Microbiology Spectrum 11.2 (2023): e02546-22. Distributed under Open Access license CC BY 4.0. https://doi.org/10.1128/spectrum.02546-22
