Lipopolysaccharide (LPS): Structure, Serotyping, and Septic Shock
Lipopolysaccharides (LPS), also known as endotoxins, are large, complex amphipathic molecules found in the outer membrane of Gram-negative bacteria. They are not merely structural components that provide physical integrity to the bacterial cell envelope; they are also potent immunomodulators capable of triggering severe immune responses in host organisms. Understanding the lipopolysaccharide structure is fundamental to microbiology and immunology, as these molecules play a dual role: they protect bacteria from environmental threats, such as antibiotics and bile salts, while simultaneously acting as the primary trigger for the septic shock mechanism in humans.
The structural diversity of LPS, particularly within the O-antigen region, forms the basis for serotyping systems used to classify bacterial strains during epidemiological outbreaks. Whether investigating the pathogenicity of Salmonella, the vaccine potential of Pseudomonas aeruginosa, or the endotoxicity of E. coli, a deep understanding of LPS biology is essential. At Creative Biolabs, we provide comprehensive Anti-Bacterial Gram-Negative Glycan (LPS) Antibody Development services to support researchers in decoding these complex glycoconjugates for therapeutic and diagnostic applications.
Structural Architecture of Lipopolysaccharide
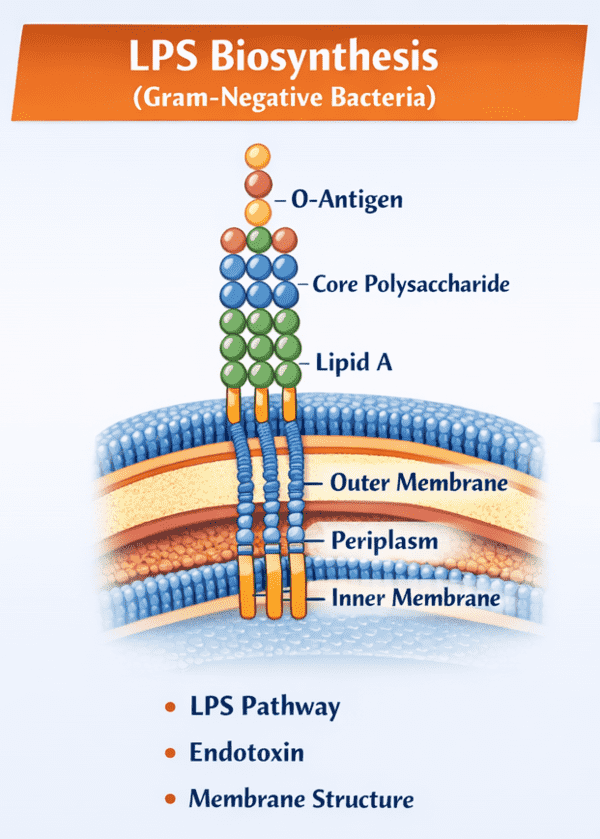
The LPS molecule is architecturally tripartite, consisting of three distinct domains covalently linked together. Each domain possesses unique chemical properties and biological functions that contribute to the overall behavior of the bacterium and its interaction with the host immune system.
1. Lipid A: The Endotoxic Center
Lipid A is the hydrophobic anchor of the LPS molecule, embedding it within the outer leaflet of the bacterial outer membrane. It is the highly conserved "bioactive" center responsible for lipid a toxicity. Structurally, it typically consists of a glucosamine disaccharide backbone that is phosphorylated and acylated with multiple fatty acid chains. The number, length, and saturation of these acyl chains vary among species but are critical for recognition by the host's innate immune receptors, specifically the Toll-like receptor 4 (TLR4) complex. It is this recognition event that initiates the potent inflammatory cascade associated with endotoxic shock.
2. Core Oligosaccharide: The Structural Bridge
Connecting Lipid A to the O-antigen is the core oligosaccharide. This domain is subdivided into the inner core and the outer core. The inner core, proximal to Lipid A, typically contains unusual sugars such as 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) and L-glycero-D-manno-heptose (Hep). These residues are vital for membrane stability and are often targets for antimicrobial peptides. The outer core consists of more common hexoses like glucose, galactose, and N-acetylglucosamine. While less variable than the O-antigen, the core structure can still exhibit variations that influence membrane permeability and susceptibility to antibiotics.
3. O-Antigen: The Variable Surface
The O-antigen (or O-polysaccharide) is the outermost domain of LPS, extending from the bacterial surface into the external environment. It is a repetitive glycan polymer comprised of oligosaccharide units (O-units) that vary extensively in sugar composition, sequence, and linkage. This extreme variability is the basis for o-antigen serotyping. For instance, in Escherichia coli alone, there are over 180 distinct O-serogroups. The O-antigen is highly immunogenic and serves as a primary target for the host's adaptive immune system. Bacteria lacking the O-antigen produce "rough" (R-form) LPS, termed lipooligosaccharides (LOS), which are often less virulent but more susceptible to host defenses.
Mechanism of Septic Shock and Immune Activation
The TLR4 Signaling Pathway
The septic shock mechanism is primarily driven by the interaction between Lipid A and the host innate immune system. When Gram-negative bacteria lyse or divide, they release LPS into the circulation. This free LPS is bound by Lipopolysaccharide-Binding Protein (LBP), which transfers it to the CD14 receptor on the surface of immune cells like macrophages and monocytes.
CD14 then presents the LPS-LBP complex to the MD-2/TLR4 receptor complex. The binding of Lipid A to MD-2 induces a conformational change in TLR4, triggering its dimerization and the recruitment of intracellular adaptor proteins such as MyD88 and TRIF. This initiates a signaling cascade that activates transcription factors like NF-κB and IRF3, leading to the massive release of pro-inflammatory cytokines (TNF-α, IL-1β, IL-6). This "cytokine storm" causes systemic vasodilation, capillary leakage, and multi-organ failure, characterizing septic shock.
O-Antigen Serotyping in Vaccine Development
Because the O-antigen is the most exposed portion of the LPS, it is a prime target for antibody-based therapeutics and vaccines. O-antigen serotyping allows researchers to categorize bacterial strains based on the unique chemical structure of their O-polysaccharide repeating units. This is critical for epidemiological tracking—identifying the specific strain responsible for an outbreak—and for designing multivalent vaccines that cover the most prevalent pathogenic serotypes. For example, current pneumococcal and meningococcal vaccines are based on similar capsular polysaccharide strategies, and next-generation vaccines against Shigella and pathogenic E. coli rely heavily on targeting specific O-antigen structures.
Our Solutions for LPS Research
The structural heterogeneity of LPS makes it a challenging molecule to study. Standard proteomic tools do not apply, and generic antibodies often fail to distinguish between closely related serotypes. Creative Biolabs offers a suite of specialized services designed to overcome these hurdles, providing high-affinity reagents and detailed structural data.
Anti-LPS Antibody Development
We generate high-specificity monoclonal and polyclonal antibodies against specific Lipid A, Core, or O-antigen epitopes. Our platform includes phage display and hybridoma technologies optimized for carbohydrate antigens, ensuring the production of antibodies that can differentiate between specific serotypes or recognize conserved core regions across species.
Microbial Glycan Antigen Microarray
Our high-throughput microarray platform allows for the simultaneous screening of antibodies or serum samples against hundreds of microbial glycan structures, including diverse LPS O-antigens. This service is essential for profiling immune responses, identifying dominant epitopes, and validating vaccine candidates.
LPS Structural Analysis
We provide comprehensive structural characterization of extracted LPS using advanced mass spectrometry (MALDI-TOF, ESI-MS), NMR spectroscopy, and gas chromatography. We can determine the precise composition of fatty acids in Lipid A, the sugar sequence of the Core, and the repeating unit structure of the O-antigen.
Custom Glycoconjugate Synthesis
For vaccine development, we offer custom conjugation services to link purified O-antigens or synthetic glycan mimics to immunogenic carrier proteins (e.g., CRM197, Tetanus Toxoid). This converts T-cell independent antigens into T-cell dependent ones, eliciting a robust and long-lasting immune memory.
Inquire About LPS Services
Published Data
A comprehensive review elucidates the sophisticated strategies employed by Gram-negative pathogens to manipulate the chemical architecture of Lipid A, thereby evading detection by the host's innate immune system. A prime example detailed in the research is Yersinia pestis, the etiological agent of plague. This pathogen exhibits a remarkable ability to remodel its LPS structure in response to environmental temperature changes associated with its life cycle. In the flea vector at 27°C, the bacterium synthesizes a hexa-acylated Lipid A, which acts as a potent agonist for the Toll-like receptor 4 (TLR4) complex. However, upon transmission to a human host at 37°C, the expression of specific acyltransferases is downregulated, shifting production to a hypo-acylated, tetra-acylated Lipid A form. This modified structure is poorly recognized by the human TLR4/MD-2 complex, effectively blinding the innate immune surveillance mechanism. This "silent" LPS phenotype prevents the induction of necessary proinflammatory cytokines, allowing the bacteria to proliferate unchecked during the early stages of infection. These insights demonstrate that Lipid A is not a static molecule but a dynamic virulence factor, emphasizing the importance of utilizing advanced structural analysis to understand pathogenesis and develop effective therapeutics against immune-evading strains.
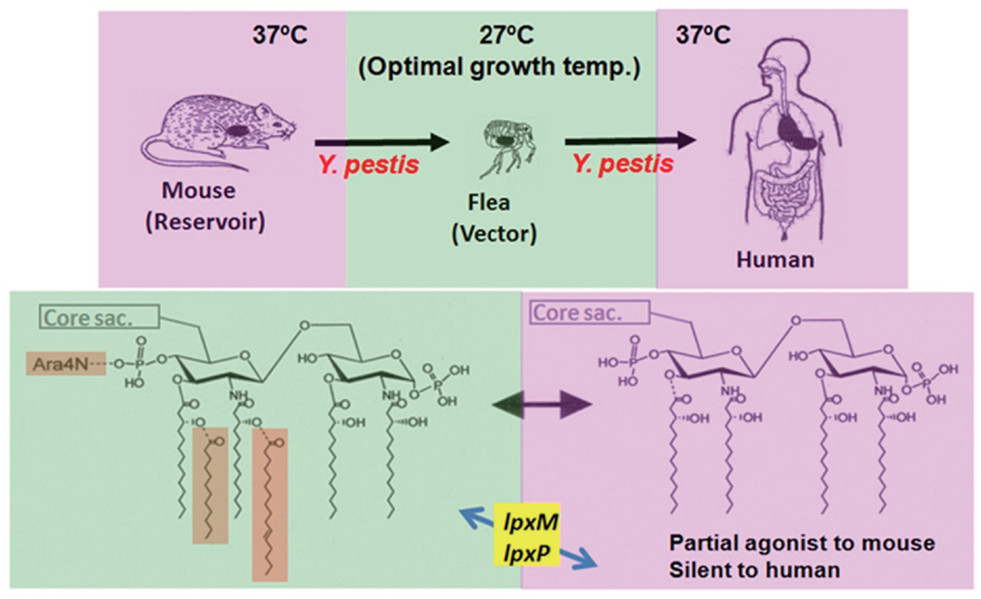 Fig.2
Temperature-dependent Lipid A structural modification in Yersinia pestis enabling host immune evasion.1
Fig.2
Temperature-dependent Lipid A structural modification in Yersinia pestis enabling host immune evasion.1
FAQs
What is the difference between LPS and LOS?
LPS (Lipopolysaccharide) typically refers to the complete molecule found in "smooth" bacterial colonies, containing Lipid A, Core, and the repetitive O-antigen. LOS (Lipooligosaccharide) refers to "rough" forms of the molecule that lack the O-antigen polysaccharide chain, consisting only of Lipid A and the core oligosaccharide. LOS is common in mucosal pathogens like Neisseria and Haemophilus.
How does Lipid A cause septic shock?
Lipid A is the bioactive endotoxin component. It binds to the TLR4/MD-2 receptor complex on host immune cells. This binding triggers an intracellular signaling cascade (via MyD88 and TRIF pathways) that activates NF-κB, leading to an uncontrolled release of pro-inflammatory cytokines (cytokine storm), resulting in systemic inflammation, vascular leakage, and organ failure.
Why is O-antigen serotyping important?
The O-antigen is highly variable among different strains of the same bacterial species. Serotyping allows epidemiologists to classify bacteria into distinct groups (serovars) based on these structural differences. This is vital for tracking the source of disease outbreaks (e.g., distinguishing pathogenic E. coli O157:H7 from commensal strains) and for designing targeted vaccines.
Can you generate antibodies against specific O-antigens?
Yes. We use specialized immunization protocols and screening strategies to generate monoclonal antibodies that recognize specific sugar residues or linkages within the O-antigen repeating unit. We can counter-screen these antibodies against related serotypes to ensure high specificity for your target strain.
What methods do you use to determine LPS structure?
We employ a combination of composition analysis (GC-MS) to identify sugar monomers and fatty acids, and advanced 1D and 2D NMR spectroscopy (COSY, TOCSY, NOESY, HSQC) to determine the sequence, linkage patterns, and anomeric configurations of the saccharide units.
Reference:
- 1. Matsuura, Motohiro. "Structural modifications of bacterial lipopolysaccharide that facilitate Gram-negative bacteria evasion of host innate immunity." Frontiers in Immunology 4 (2013): 109. Distributed under Open Access license CC BY 3.0. https://doi.org/10.3389/fimmu.2013.00109
