Structure based Screening
Structure-based screening refers to the emerging technology by molecular composite docking, based on the three-dimensional structure of the receptor, which can automatically match small molecules in the compound database at the binding site and predict its binding mode to obtain a composite energy ranking. In recent years, virtual screening methods in computer-aided drug design have become a reliable and relatively inexpensive technique for discovering lead compounds. Creative Biolabs is committed to creating a pre-clinical technical service platform with rich screening models, experimental science, rigorous operation, and credible results, especially in the field of structure-based virtual screening, and strives to provide high-quality and efficient scientific and technical services for global drug research and development users.
Ligand-docking Structure-based Screening
The development of ligand-based prediction methods originated in the late 1970s and continues to the present. The prediction based on ligand structure refers to a method based on drug design theory, using computer technology and professional application software to select some compounds from a large number of compounds even a million molecules, and to evaluate the activity of the experiment. This approach requires first determining the ligand-receptor structure rather than the pre-existing ligand, so the method can support a variety of customization needs of the customer.
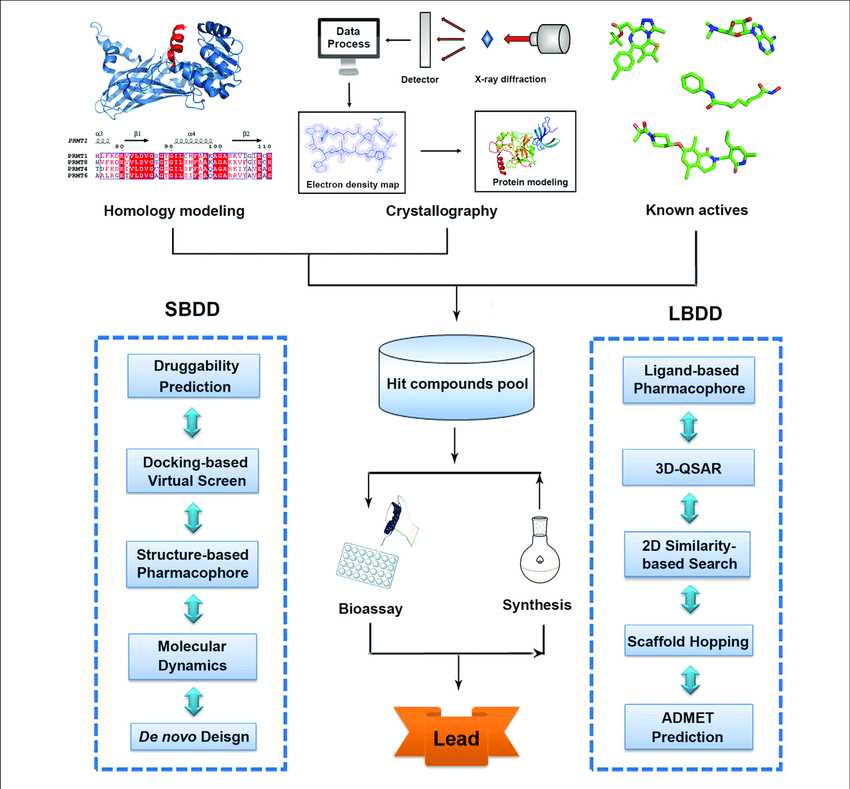 Fig.1 Traditional workflow of structure-based drug design (SBDD) and ligand-based drug design (LBDD). (Lu, 2018)
Fig.1 Traditional workflow of structure-based drug design (SBDD) and ligand-based drug design (LBDD). (Lu, 2018)
Fragment-based De Novo Methods
Fragment-based de novo methods were inspired by research in the early 1990s, and scientists have found that a biologically active drug molecule can be viewed as the result of a superposition of biological activity of two or more small molecule fragments. Usually, a known drug molecule is tailored into multiple fragments, some of which may inherit all or part of the pharmacological properties of the original active molecule, and by screening these molecular fragments, it is possible to find a better drug molecule. The advantage of this technology is that scientists can find lead compounds with higher biological activity in a shorter time without a lot of manpower and material resources.
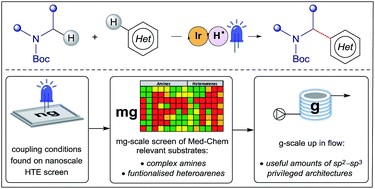 Fig.2 Schematic diagram of fragment-based drug synthesis. (Grainger, 2019)
Fig.2 Schematic diagram of fragment-based drug synthesis. (Grainger, 2019)
Protocol for Structure-based Screening
The basic steps of structure-based screening can be summarized as follows:
- Design targeted targets, one or more of these targeted targets will be designed. Often these targets are closely related to a particular disease, and inhibiting or altering these targets will help treat the disease.
- Design and synthesis of a library of compounds containing a variety of small-molecule fragments of drugs. The ideal small molecule fragment should have a certain affinity for the active site of the target, such as the target protein or the targeting enzyme.
- Screening of small fragments of synthesized fragments using pre-designed targeted targets.
- According to the results of the screening, molecular fragments with better activity can be obtained, and the lead compounds can be obtained by appropriately combining the fragments.
- The chemical structure of the lead compound that has been obtained is further modified and further optimized to obtain a drug candidate for clinical research.
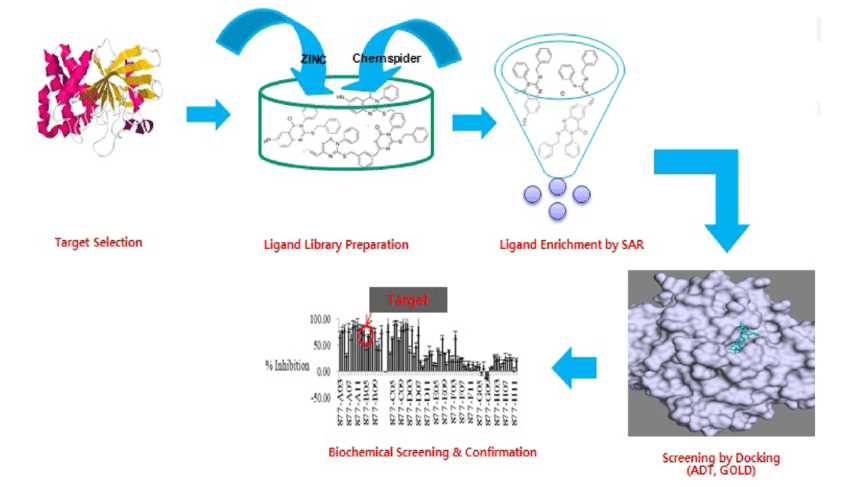 Fig.3 A conceptual figure of structure-based virtual screening, illustrating the schematic design and typical drug discovery steps in the search for a potential protein tyrosine phosphatase inhibitor. (Reddy, 2017)
Fig.3 A conceptual figure of structure-based virtual screening, illustrating the schematic design and typical drug discovery steps in the search for a potential protein tyrosine phosphatase inhibitor. (Reddy, 2017)
Creative Biolabs owns many high standard labs which are equipped with molecular fragmentation and protein crystallography-based drug discovery and screening platforms to support structurally based drug development, from confirmation of new targets to final structural validation. If you are interested in the structure-based screening, please contact us and we will happy to help you.
References
- Lu, W.; et al. Computer-aided drug design in epigenetics. Frontiers in Chemistry. 2018, 6: 57.
- Grainger, R.; et al. Enabling synthesis in fragment-based drug discovery by reactivity mapping: photoredox-mediated cross-dehydrogenative heteroarylation of cyclic amines. Chemical Science. 2019, 10(8): 2264-2271.
- Reddy, R.H.; et al. Structure-based virtual screening of protein tyrosine phosphatase inhibitors: significance, challenges and solutions. Journal of Microbiology and Biotechnology. 2017, 27(5): 878-895.
For Research Use Only.
