Virtual Combinatorial Library
Creative Biolabs is a world-leading service provider who is committing to delivering a full range of drug discovery services for global clients with high quality and low cost. With rich experience in virtual combinatorial library design, we are confident in offering the best services for identification of drug lead candidates.
Virtual combinatorial library is made of all possible combinations in number and nature of the available components. It allows selection of the best binding constituent from all available candidates, accelerating the chances of identifying leads compounds for drug discovery. Virtual combinatorial library has been regarded as an established research tool for the discovery of substrates, inhibitors, receptors, catalysts, and carriers for a variety of processes.
Design of Virtual Combinatorial Libraries
Virtual combinatorial library is a powerful methodology for the discovery of drug. By virtual combinatorial libraries, novel chemical compounds can be obtained, while they are not available to be obtained when screening existing compound libraries. Virtual combinatorial library design integrates all methods that have been developed for the virtual screening of existing compound libraries. The major methods include similarity-based compound clustering techniques and structure-based docking and scoring. Traditionally, combinatorial libraries have been enumerated from a set of predefined building blocks and filtered using predictive and filtering software. However, the number of structures virtually assembled may be limited by the special software used and computer hardware. The trend in virtual combinatorial library design is to integrate the library generation process (design) and the evaluation procedure (scoring). In other words, the combinatorial library construction is guided by predictions and the degrees of structural freedom defined by chemistry.
Strategies for Virtual Combinatorial Library Design
- Ligand Structure-Based Design
When there is little or no structural information available about the target, ligand structure-based methods represent available approaches to combinatorial library design. Typically, the starting point for ligand-based library design is the calculation of descriptors to characterize target molecular structures, followed by quantitative structure-activity relationship (QSAR) to optimize for the desired library characteristics. Currently, the most successful methodologies are based on a pharmacophore hypothesis. Isofunctional molecular architectures can be virtually assembled mimicking the pharmacophore pattern present in the original template based on a pharmacophore model that was derived from a set of known bioactive molecules or ligands.
- Protein Structure-Based Design
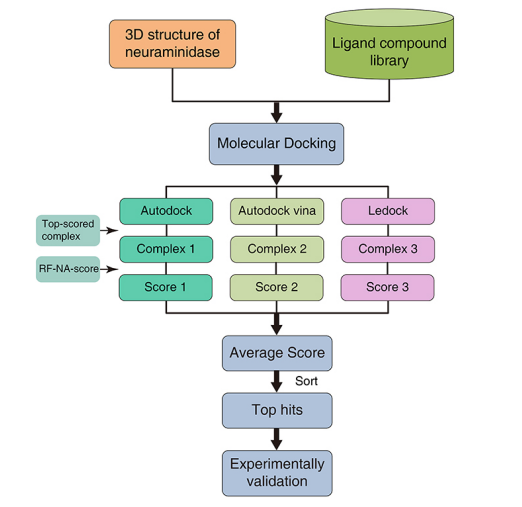 Fig.1 Virtual screening strategy using molecular docking software and RF-NA-Score. (Zhang, 2017)
Fig.1 Virtual screening strategy using molecular docking software and RF-NA-Score. (Zhang, 2017)
It has become an essential part of structure-based drug discovery that utilizes rapid docking and scoring routines to screen a list of virtual ligands. Docking methods are used to find a match between each ligand and a set of site points and then scoring functions are applied to estimate binding energy. In general, a better prediction of binding modes can be obtained by the combined use of docking together with pharmacophoric restraints and knowledge-based consensus, increasing enrichment factors through novel “hybrid” scoring functions.
- Interaction Based Design
Interaction based design is a new design method developed in recent years. It integrates the power of ligand and protein-based design with experimental protein-ligand data.
What Can We Do?
Creative Biolabs has accumulated a lot of experience in drug discovery using virtual screening techniques. Our excellent scientists can customize a variety of project alternatives from which you can find the best for you. We are committed to offering the top-ranking and comprehensive services for virtual combinatorial libraries design, accelerating the process of drug discovery. For more information and inquiries about Creative Biolabs' virtual combinatorial library design services, please do not hesitate to contact us.
Reference
- Zhang, L.; et al. Virtual screening approach to identifying influenza virus neuraminidase inhibitors using molecular docking combined with machine-learning-based scoring function. Oncotarget. 2017, 8: 83142-83154.
For Research Use Only.
